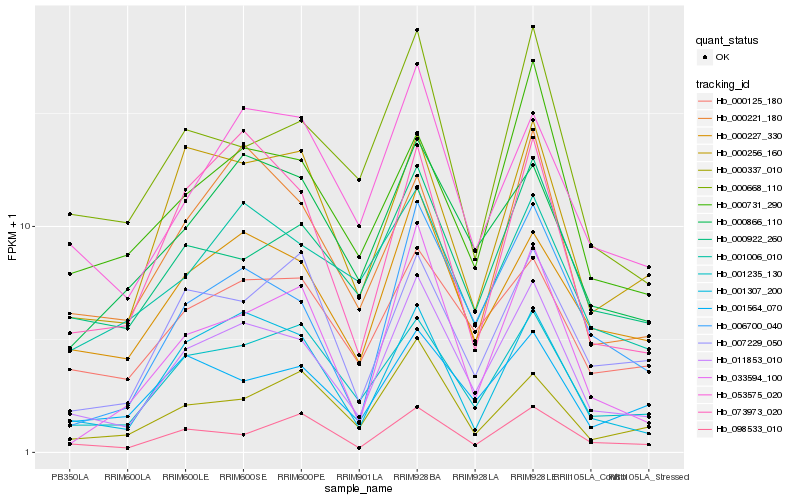
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011853_010 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 2 |
Hb_006700_040 |
0.1143388226 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 3 |
Hb_000227_330 |
0.115533009 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 4 |
Hb_001235_130 |
0.1205816545 |
- |
- |
- |
| 5 |
Hb_001307_200 |
0.1221822819 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 6 |
Hb_000922_260 |
0.1303198415 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 7 |
Hb_000125_180 |
0.1318422962 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 8 |
Hb_053575_020 |
0.1330361012 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 9 |
Hb_000256_160 |
0.1354270958 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 10 |
Hb_033594_100 |
0.1359271959 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000668_110 |
0.1373997198 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 12 |
Hb_098533_010 |
0.1380759825 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 13 |
Hb_000866_110 |
0.1404577695 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 14 |
Hb_073973_020 |
0.1407634024 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 15 |
Hb_001564_070 |
0.1420316051 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 16 |
Hb_000337_010 |
0.1457313877 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000731_290 |
0.147813005 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000221_180 |
0.1478358764 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 19 |
Hb_007229_050 |
0.1482422966 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001006_010 |
0.1484432211 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |