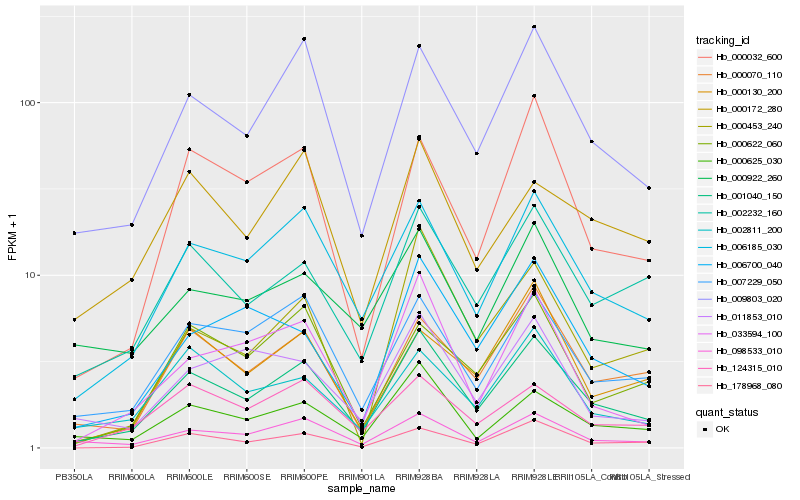
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006185_030 |
0.0945148571 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 3 |
Hb_009803_020 |
0.0999324854 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 4 |
Hb_098533_010 |
0.1159522712 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 5 |
Hb_007229_050 |
0.1228395306 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_178968_080 |
0.1228672691 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000625_030 |
0.1252180543 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 8 |
Hb_000070_110 |
0.1263564973 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002232_160 |
0.127136396 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 10 |
Hb_000032_600 |
0.1334113577 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_124315_010 |
0.1350865343 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 12 |
Hb_033594_100 |
0.1391142312 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002811_200 |
0.1447827685 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 14 |
Hb_006700_040 |
0.1450208682 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 15 |
Hb_000622_060 |
0.1501332189 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000172_280 |
0.1526724609 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_000922_260 |
0.1530771734 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 18 |
Hb_000453_240 |
0.1533638472 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 19 |
Hb_011853_010 |
0.1544285662 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 20 |
Hb_000130_200 |
0.1547377096 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |