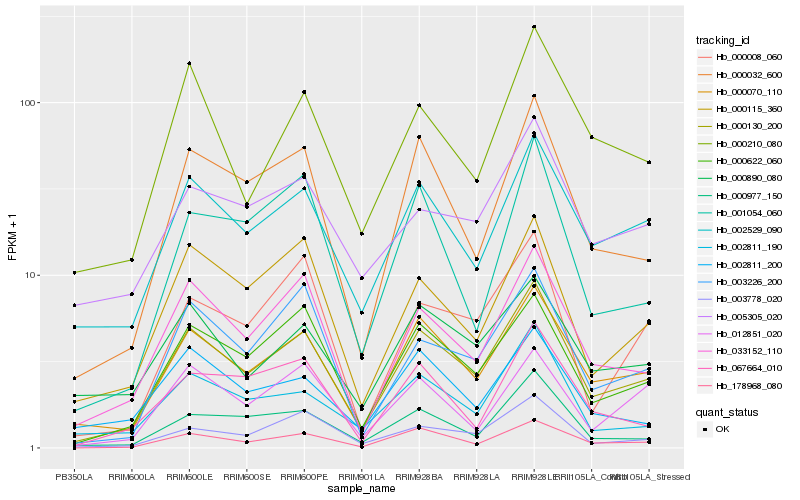
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_200 |
0.0 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 2 |
Hb_000070_110 |
0.0731646389 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000622_060 |
0.0937552904 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_178968_080 |
0.0978033162 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000032_600 |
0.1006393522 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 6 |
Hb_033152_110 |
0.1010426222 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 7 |
Hb_002529_090 |
0.1080503773 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_000115_360 |
0.1206358257 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 9 |
Hb_003226_200 |
0.1209509289 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 10 |
Hb_003778_020 |
0.12219629 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000977_150 |
0.1298027553 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 12 |
Hb_000210_080 |
0.1318796368 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_002811_190 |
0.1331107247 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 14 |
Hb_000008_060 |
0.1388292518 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000890_080 |
0.1389244739 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 16 |
Hb_001054_060 |
0.1449251883 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 17 |
Hb_002811_200 |
0.1454927474 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 18 |
Hb_067664_010 |
0.1461694793 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 19 |
Hb_012851_020 |
0.1480583095 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_005305_020 |
0.148515793 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |