| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
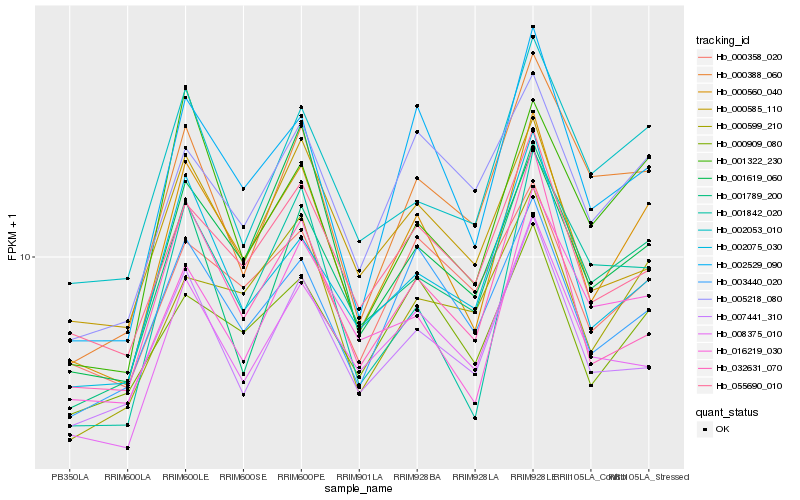
Hb_000560_040 |
0.0 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002529_090 |
0.0881898332 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_055690_010 |
0.103368095 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003440_020 |
0.10464309 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002075_030 |
0.1076295325 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_032631_070 |
0.1088431429 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_002053_010 |
0.1094864404 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000909_080 |
0.1117560732 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 9 |
Hb_016219_030 |
0.1128843477 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_001619_060 |
0.1132484249 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001789_200 |
0.1133707203 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008375_010 |
0.1152973468 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 13 |
Hb_005218_080 |
0.1153755037 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001322_230 |
0.1155986744 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 15 |
Hb_000599_210 |
0.1157478348 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 16 |
Hb_007441_310 |
0.1164057288 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000585_110 |
0.1174415917 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 18 |
Hb_000358_020 |
0.1178667981 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 19 |
Hb_001842_020 |
0.1186634404 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 20 |
Hb_000388_060 |
0.119543359 |
- |
- |
fructokinase [Manihot esculenta] |