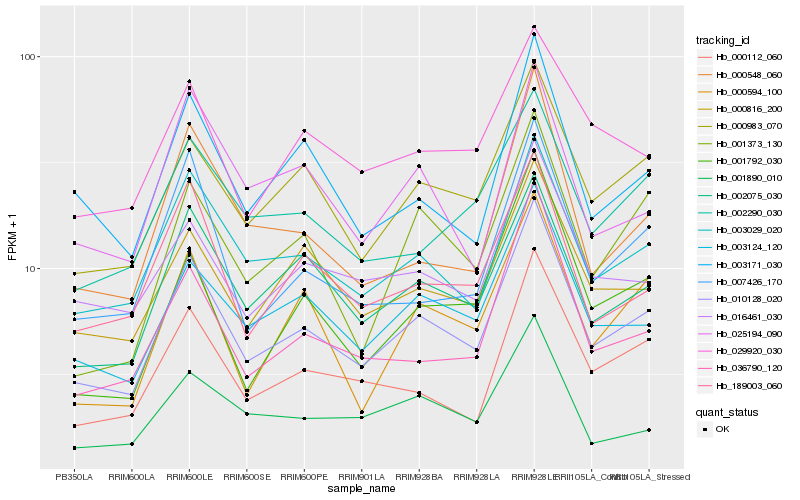
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010128_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 2 |
Hb_003029_020 |
0.0787646755 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 3 |
Hb_003124_120 |
0.0795582689 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 4 |
Hb_000983_070 |
0.0813561048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002075_030 |
0.0847424897 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000816_200 |
0.0955187388 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_000112_060 |
0.0979070939 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 8 |
Hb_016461_030 |
0.1000274473 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_003171_030 |
0.1020742686 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 10 |
Hb_189003_060 |
0.102104646 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_036790_120 |
0.1034122927 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000548_060 |
0.1072051664 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 13 |
Hb_007426_170 |
0.1083464555 |
transcription factor |
TF Family: TCP |
- |
| 14 |
Hb_002290_030 |
0.1100119025 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000594_100 |
0.1108952992 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 16 |
Hb_029920_030 |
0.11201199 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001890_010 |
0.1121835318 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_001373_130 |
0.1133785824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001792_030 |
0.1153487191 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 20 |
Hb_025194_090 |
0.1167282633 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |