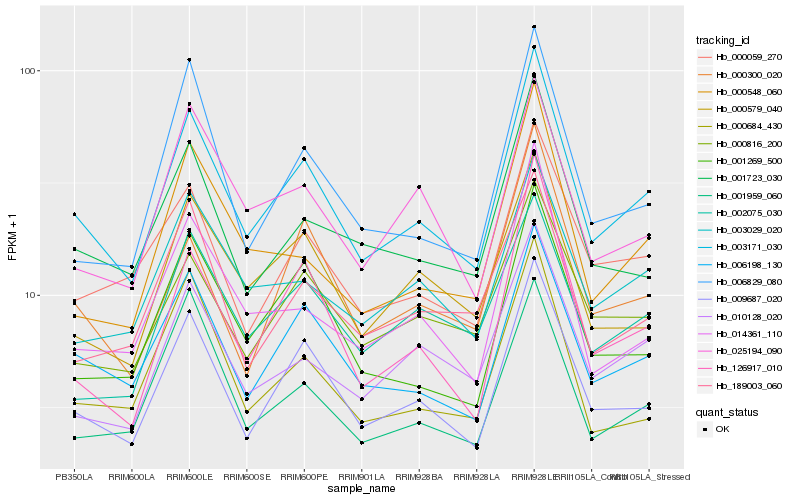
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003171_030 |
0.0 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 2 |
Hb_009687_020 |
0.0649238926 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 3 |
Hb_006198_130 |
0.0706499773 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 4 |
Hb_000059_270 |
0.0949316017 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 5 |
Hb_000816_200 |
0.0993115043 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 6 |
Hb_001723_030 |
0.0998694931 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001269_500 |
0.1014747554 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 8 |
Hb_010128_020 |
0.1020742686 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 9 |
Hb_003029_020 |
0.1026638036 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 10 |
Hb_000579_040 |
0.1056447862 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 11 |
Hb_126917_010 |
0.1060828573 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 12 |
Hb_006829_080 |
0.1071120004 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 13 |
Hb_000300_020 |
0.1073371315 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 14 |
Hb_025194_090 |
0.1087299152 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 15 |
Hb_000548_060 |
0.1089373767 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 16 |
Hb_014361_110 |
0.109976551 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001959_060 |
0.1103829799 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002075_030 |
0.1124439951 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000684_430 |
0.1127061301 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 20 |
Hb_189003_060 |
0.1128872313 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |