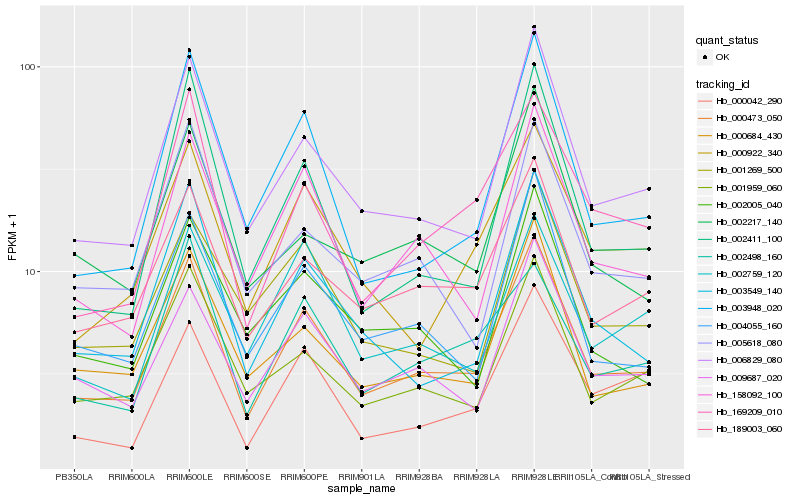
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000473_050 |
0.0 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 2 |
Hb_006829_080 |
0.0741040074 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 3 |
Hb_189003_060 |
0.0851732977 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_158092_100 |
0.0924934744 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 5 |
Hb_009687_020 |
0.0927940639 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 6 |
Hb_001959_060 |
0.0976647263 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000684_430 |
0.1027193021 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004055_160 |
0.1031464959 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_169209_010 |
0.1041459712 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003549_140 |
0.1082382867 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_003948_020 |
0.1090492832 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 12 |
Hb_000922_340 |
0.1095594919 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 13 |
Hb_000042_290 |
0.1128832798 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002498_160 |
0.1135810184 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 15 |
Hb_001269_500 |
0.1165834556 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 16 |
Hb_005618_080 |
0.1181235845 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002759_120 |
0.1193637591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002217_140 |
0.1205974305 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 19 |
Hb_002411_100 |
0.121450835 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 20 |
Hb_002005_040 |
0.1216962921 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |