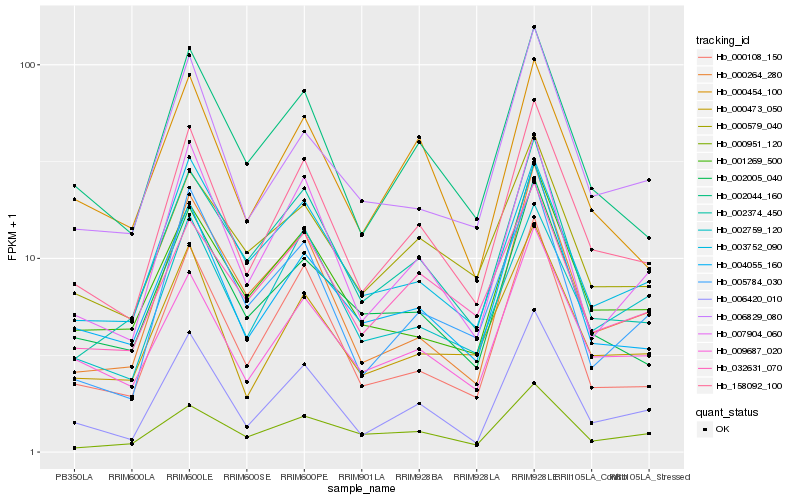
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158092_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 2 |
Hb_002044_160 |
0.0776342982 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 3 |
Hb_009687_020 |
0.0820042182 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 4 |
Hb_006420_010 |
0.0834902613 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 5 |
Hb_007904_060 |
0.0874661368 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 6 |
Hb_000473_050 |
0.0924934744 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 7 |
Hb_004055_160 |
0.0944245164 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000108_150 |
0.0958129006 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 9 |
Hb_002759_120 |
0.0999981201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002005_040 |
0.1000166211 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_002374_450 |
0.1010750173 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001269_500 |
0.1044810248 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 13 |
Hb_000264_280 |
0.1053260516 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 14 |
Hb_003752_090 |
0.1054937861 |
- |
- |
chitinase, putative [Ricinus communis] |
| 15 |
Hb_000454_100 |
0.1057201516 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_006829_080 |
0.106197283 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 17 |
Hb_032631_070 |
0.1080501601 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000579_040 |
0.108405448 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 19 |
Hb_000951_120 |
0.1134601721 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 20 |
Hb_005784_030 |
0.1143811284 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |