| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
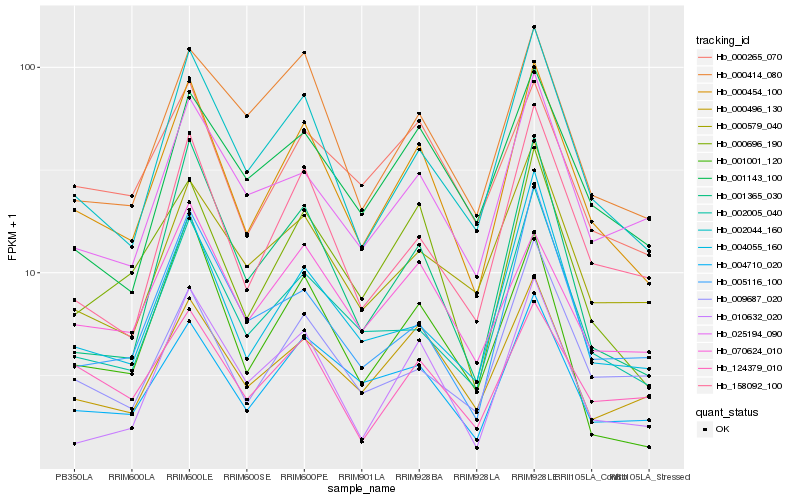
Hb_000454_100 |
0.0 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 2 |
Hb_002044_160 |
0.0810582032 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 3 |
Hb_000696_190 |
0.0972489086 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 4 |
Hb_004710_020 |
0.102777051 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 5 |
Hb_158092_100 |
0.1057201516 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 6 |
Hb_002005_040 |
0.1109479477 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000496_130 |
0.1129735956 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 8 |
Hb_010632_020 |
0.1151809966 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 9 |
Hb_004055_160 |
0.1204777824 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001365_030 |
0.1219060915 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 11 |
Hb_025194_090 |
0.1233839293 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 12 |
Hb_000414_080 |
0.1234166835 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000265_070 |
0.1237681681 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 14 |
Hb_000579_040 |
0.125481222 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 15 |
Hb_070624_010 |
0.1256216621 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 16 |
Hb_001143_100 |
0.1265324696 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 17 |
Hb_009687_020 |
0.1270220833 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 18 |
Hb_124379_010 |
0.1283034547 |
- |
- |
PREDICTED: notchless protein homolog [Citrus sinensis] |
| 19 |
Hb_005116_100 |
0.1289060618 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 20 |
Hb_001001_120 |
0.1289348254 |
- |
- |
protein phosphatase, putative [Ricinus communis] |