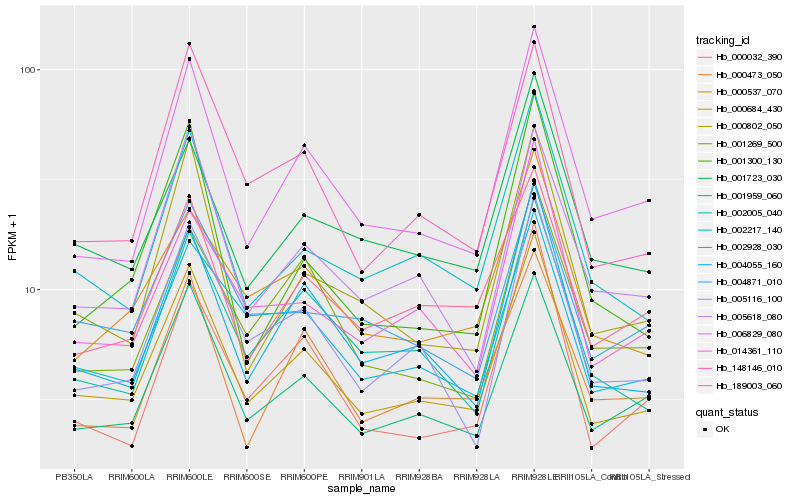
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_430 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001959_060 |
0.0723177696 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004055_160 |
0.0744007651 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002217_140 |
0.07606353 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 5 |
Hb_006829_080 |
0.0791571038 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 6 |
Hb_148146_010 |
0.0857986311 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 7 |
Hb_002005_040 |
0.0929928259 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_001723_030 |
0.0947275504 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005116_100 |
0.0959759861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 10 |
Hb_000802_050 |
0.0970174318 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 11 |
Hb_000537_070 |
0.0972587704 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 12 |
Hb_005618_080 |
0.0972627632 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_014361_110 |
0.0973806073 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002928_030 |
0.097660912 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 15 |
Hb_189003_060 |
0.0997266601 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000473_050 |
0.1027193021 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 17 |
Hb_000032_390 |
0.1067129677 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 18 |
Hb_004871_010 |
0.1068883645 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 19 |
Hb_001300_130 |
0.1084494955 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 20 |
Hb_001269_500 |
0.1102163009 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |