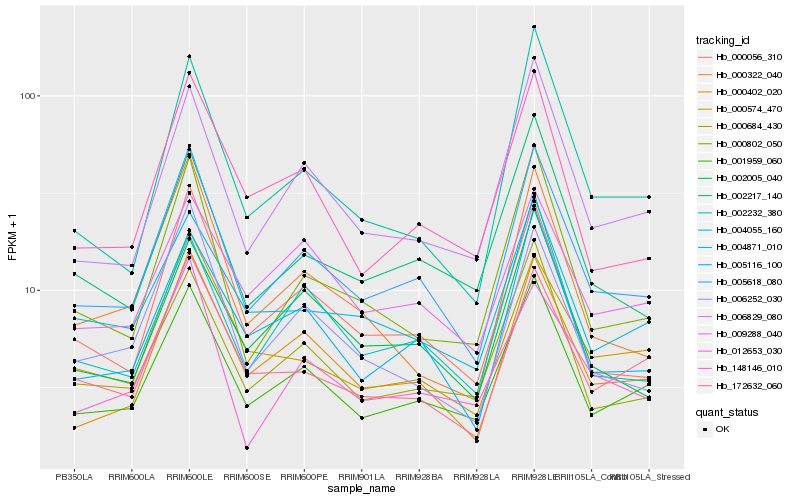
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005618_080 |
0.0 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001959_060 |
0.0700560785 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000402_020 |
0.0751996034 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_005116_100 |
0.0861692641 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 5 |
Hb_000056_310 |
0.0863950421 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 6 |
Hb_006829_080 |
0.0894351702 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 7 |
Hb_172632_060 |
0.0968376476 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000684_430 |
0.0972627632 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000802_050 |
0.0975880764 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 10 |
Hb_148146_010 |
0.1017742231 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 11 |
Hb_006252_030 |
0.1018633344 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 12 |
Hb_002005_040 |
0.1036918491 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000574_470 |
0.104937135 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002217_140 |
0.1050087456 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 15 |
Hb_002232_380 |
0.1075534267 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_012653_030 |
0.1081340825 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_004055_160 |
0.1092619828 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004871_010 |
0.1098629474 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 19 |
Hb_009288_040 |
0.113216782 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 20 |
Hb_000322_040 |
0.1143113159 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |