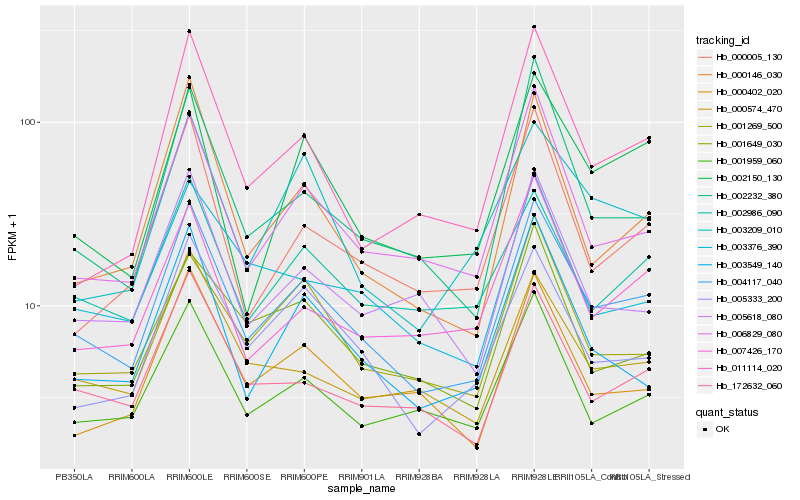
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 2 |
Hb_006829_080 |
0.1053677036 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 3 |
Hb_172632_060 |
0.1084604162 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001959_060 |
0.1103791098 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005333_200 |
0.116322248 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002150_130 |
0.1169367168 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002986_090 |
0.1172633631 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_000146_030 |
0.1179633204 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000574_470 |
0.1192281672 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_002232_380 |
0.1201154527 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000402_020 |
0.1216043949 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_005618_080 |
0.1217391517 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003376_390 |
0.1224370618 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003549_140 |
0.1224647789 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003209_010 |
0.1240487144 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 16 |
Hb_000005_130 |
0.1247890182 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 17 |
Hb_011114_020 |
0.1253957184 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001269_500 |
0.1255476881 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 19 |
Hb_001649_030 |
0.1261148469 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007426_170 |
0.1264429258 |
transcription factor |
TF Family: TCP |
- |