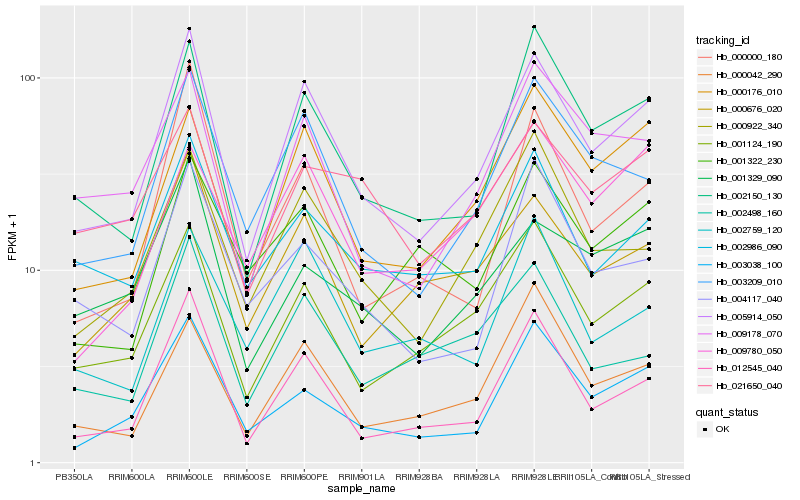
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005914_050 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002150_130 |
0.0997677045 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003209_010 |
0.1045643174 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 4 |
Hb_012545_040 |
0.1084417023 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001124_190 |
0.1108391578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000176_010 |
0.1146087037 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000676_020 |
0.1211316122 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 8 |
Hb_003038_100 |
0.1244890045 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000922_340 |
0.125229523 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 10 |
Hb_009178_070 |
0.1277448458 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002986_090 |
0.1328696891 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_000042_290 |
0.1359484433 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002759_120 |
0.1418894375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002498_160 |
0.1421410316 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 15 |
Hb_000000_180 |
0.142223836 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_004117_040 |
0.1433164076 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 17 |
Hb_001329_090 |
0.1438837657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_021650_040 |
0.1507031697 |
- |
- |
EG2771 [Manihot esculenta] |
| 19 |
Hb_001322_230 |
0.1517314318 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 20 |
Hb_009780_050 |
0.1528636708 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |