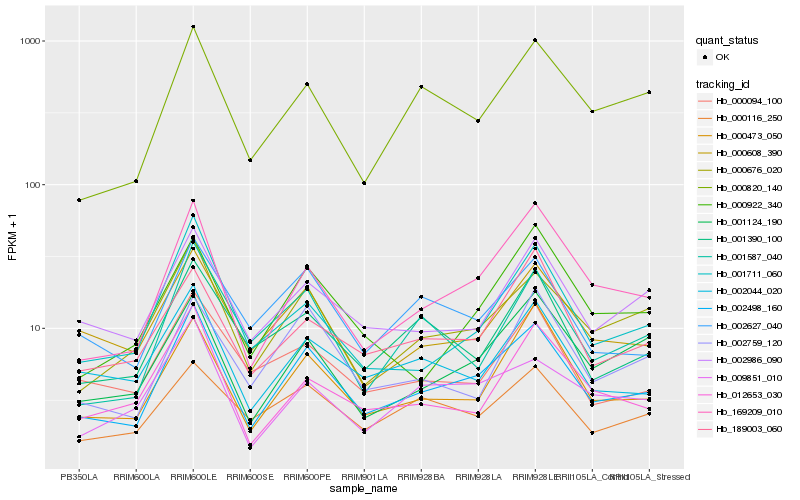
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 2 |
Hb_169209_010 |
0.1004842358 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000676_020 |
0.1067116784 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 4 |
Hb_000473_050 |
0.1135810184 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 5 |
Hb_000608_390 |
0.116739052 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 6 |
Hb_002044_020 |
0.1218645886 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 7 |
Hb_002986_090 |
0.1258433525 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_001124_190 |
0.1260239653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002627_040 |
0.1280757196 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 10 |
Hb_001711_060 |
0.1289152224 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 11 |
Hb_000820_140 |
0.1293288287 |
- |
- |
histone H4 [Zea mays] |
| 12 |
Hb_001390_100 |
0.1339953701 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009851_010 |
0.1346635954 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000094_100 |
0.1346689287 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 15 |
Hb_189003_060 |
0.1356475025 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_012653_030 |
0.1364600595 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_000922_340 |
0.1379919906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 18 |
Hb_001587_040 |
0.1394302434 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002759_120 |
0.1410578343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000116_250 |
0.1414036563 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |