| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
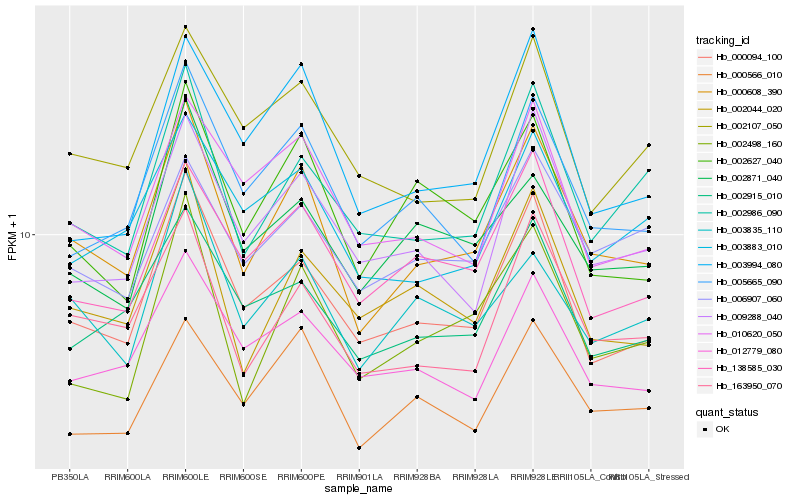
Hb_000608_390 |
0.0 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 2 |
Hb_163950_070 |
0.088699685 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 3 |
Hb_000094_100 |
0.0913038917 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 4 |
Hb_005665_090 |
0.1049508543 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 5 |
Hb_002986_090 |
0.1083722436 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_002044_020 |
0.1106702206 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 7 |
Hb_002915_010 |
0.1134693026 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 8 |
Hb_002627_040 |
0.1136902042 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_012779_080 |
0.114751844 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_003835_110 |
0.1156335832 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_138585_030 |
0.1158494172 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_009288_040 |
0.116015823 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 13 |
Hb_002498_160 |
0.116739052 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 14 |
Hb_010620_050 |
0.1176838777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 15 |
Hb_003883_010 |
0.1205395636 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 16 |
Hb_002107_050 |
0.1220511187 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 17 |
Hb_000566_010 |
0.1224360762 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_002871_040 |
0.1238185686 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 19 |
Hb_003994_080 |
0.1243233521 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 20 |
Hb_006907_060 |
0.1246449282 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |