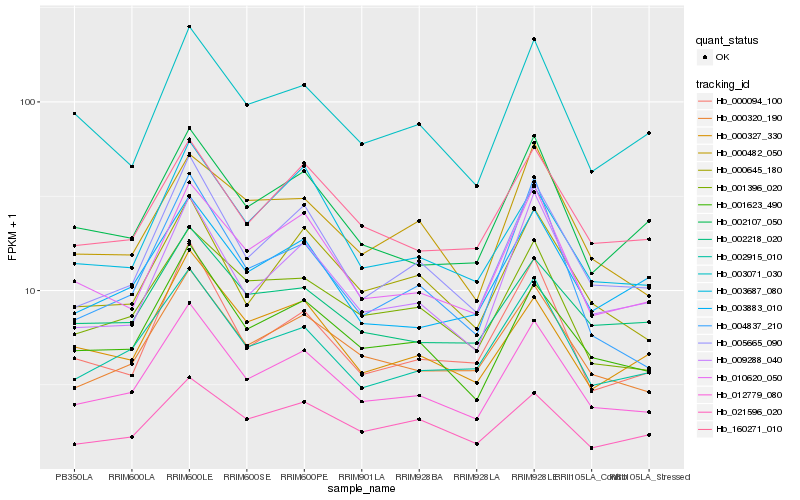
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012779_080 |
0.0 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 2 |
Hb_005665_090 |
0.0518747735 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000320_190 |
0.0546403414 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001623_490 |
0.0583468092 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002915_010 |
0.0652644832 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 6 |
Hb_010620_050 |
0.0677628112 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 7 |
Hb_009288_040 |
0.0762527146 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 8 |
Hb_000645_180 |
0.0765105139 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_002107_050 |
0.0790819898 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 10 |
Hb_001396_020 |
0.0791710524 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 11 |
Hb_000094_100 |
0.0796678207 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 12 |
Hb_160271_010 |
0.0797120484 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_003071_030 |
0.0843761451 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003883_010 |
0.0859998291 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 15 |
Hb_002218_020 |
0.0880594992 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 16 |
Hb_000327_330 |
0.0894587944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003687_080 |
0.0901866079 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 18 |
Hb_000482_050 |
0.0908167899 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_021596_020 |
0.0922677954 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 20 |
Hb_004837_210 |
0.0935010405 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |