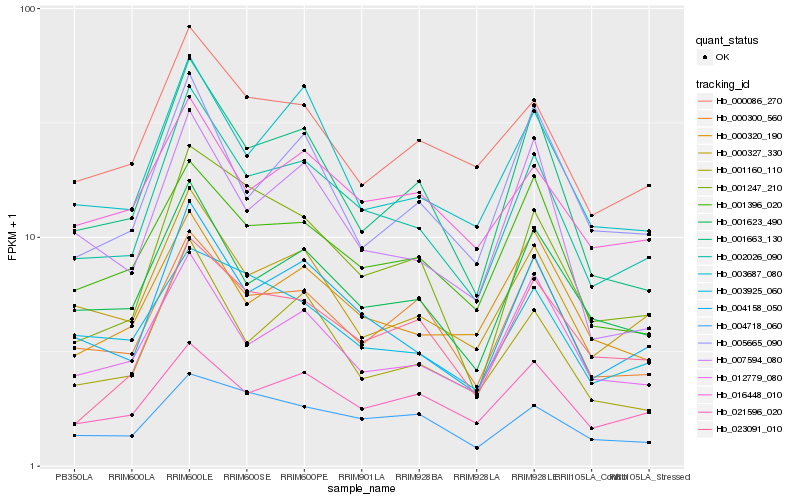
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_090 |
0.0 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 2 |
Hb_004158_050 |
0.0531806188 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 3 |
Hb_001623_490 |
0.0805192589 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000327_330 |
0.0859549953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001160_110 |
0.0900302738 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 6 |
Hb_001663_130 |
0.0905832144 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 7 |
Hb_003687_080 |
0.0953861825 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 8 |
Hb_000300_560 |
0.0972667029 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012779_080 |
0.1014494115 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001396_020 |
0.1015126373 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 11 |
Hb_000086_270 |
0.1020111629 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 12 |
Hb_021596_020 |
0.1030180477 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 13 |
Hb_016448_010 |
0.1031161106 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 14 |
Hb_004718_060 |
0.1045450975 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001247_210 |
0.1058865215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003925_060 |
0.1105728651 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000320_190 |
0.1117420317 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_005665_090 |
0.1119684389 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 19 |
Hb_007594_080 |
0.1125024648 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 20 |
Hb_023091_010 |
0.1128481668 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |