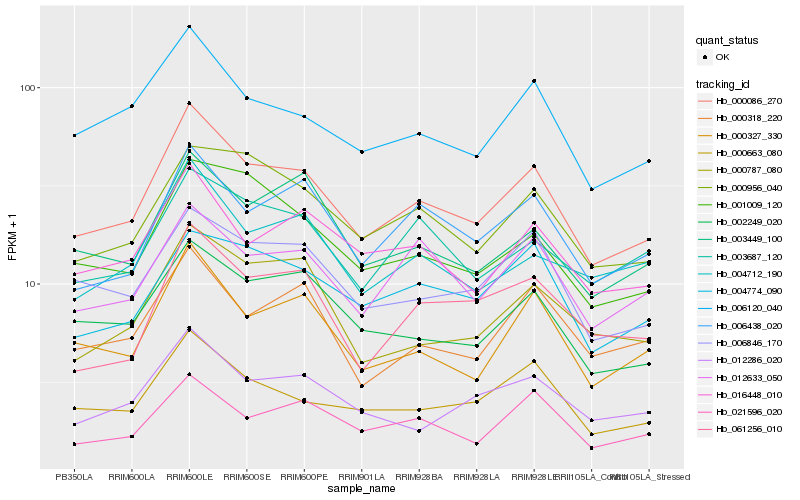
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_270 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 2 |
Hb_006120_040 |
0.0798287597 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 3 |
Hb_000327_330 |
0.0803776817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002249_020 |
0.0838650509 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 5 |
Hb_000663_080 |
0.084221703 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 6 |
Hb_006438_020 |
0.0855730552 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 7 |
Hb_016448_010 |
0.0862230329 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 8 |
Hb_006846_170 |
0.0865420446 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 9 |
Hb_000956_040 |
0.0893539321 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 10 |
Hb_021596_020 |
0.0921788392 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 11 |
Hb_001009_120 |
0.0929176209 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004712_190 |
0.0931857601 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 13 |
Hb_012633_050 |
0.0934674414 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000318_220 |
0.0941331094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003449_100 |
0.0942206047 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 16 |
Hb_012286_020 |
0.0955335518 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 17 |
Hb_000787_080 |
0.0962891089 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 18 |
Hb_004774_090 |
0.0968135267 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 19 |
Hb_061256_010 |
0.0971563901 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003687_120 |
0.0980808894 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |