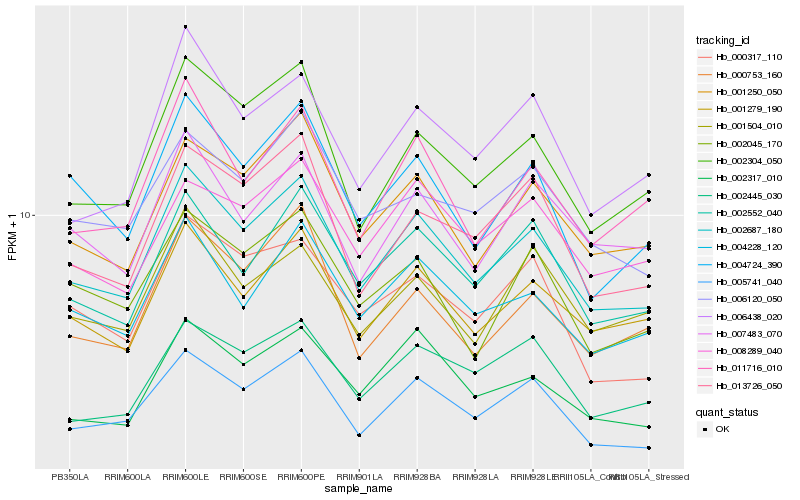
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_180 |
0.0 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 2 |
Hb_001279_190 |
0.0552807829 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004228_120 |
0.0600385054 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 4 |
Hb_001250_050 |
0.0612305984 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002304_050 |
0.0617005785 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 6 |
Hb_002552_040 |
0.0621446083 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 7 |
Hb_005741_040 |
0.0668931033 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 8 |
Hb_011716_010 |
0.067907555 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 9 |
Hb_004724_390 |
0.0708536165 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 10 |
Hb_013726_050 |
0.0731071897 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 11 |
Hb_006438_020 |
0.074152861 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 12 |
Hb_007483_070 |
0.0747790999 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 13 |
Hb_006120_050 |
0.0753509915 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 14 |
Hb_002317_010 |
0.076569559 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_008289_040 |
0.0771730799 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 16 |
Hb_002045_170 |
0.077974802 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000317_110 |
0.0785675312 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 18 |
Hb_001504_010 |
0.0806069419 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 19 |
Hb_002445_030 |
0.0812622743 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 20 |
Hb_000753_160 |
0.0812775014 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |