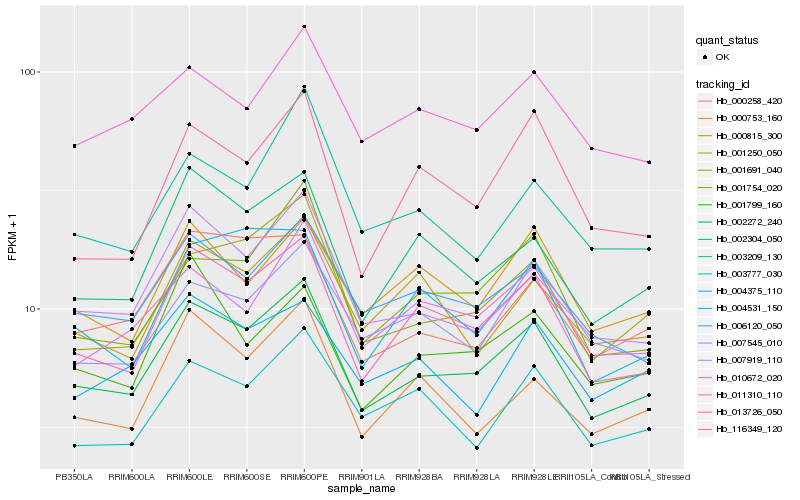
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002272_240 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_013726_050 |
0.0528730629 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 3 |
Hb_001691_040 |
0.0728050693 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007919_110 |
0.074790795 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 5 |
Hb_006120_050 |
0.0750308364 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 6 |
Hb_002304_050 |
0.0783900716 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 7 |
Hb_001754_020 |
0.0833327568 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 8 |
Hb_010672_020 |
0.083663956 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000258_420 |
0.0841723681 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 10 |
Hb_011310_110 |
0.0860351467 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000815_300 |
0.090664439 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 12 |
Hb_003777_030 |
0.091735907 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 13 |
Hb_000753_160 |
0.0925085189 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_001250_050 |
0.0927332185 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007545_010 |
0.0939922461 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 16 |
Hb_003209_130 |
0.0941966079 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_004375_110 |
0.0949428671 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 18 |
Hb_116349_120 |
0.0951248373 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_001799_160 |
0.0957000081 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_004531_150 |
0.0964936558 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |