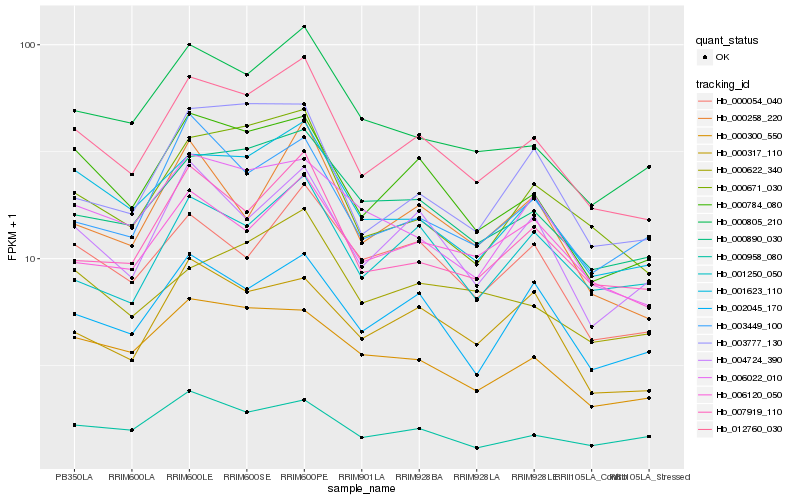
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_030 |
0.0 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 2 |
Hb_000784_080 |
0.0677121518 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 3 |
Hb_001623_110 |
0.0687463161 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_000890_030 |
0.0737457564 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 5 |
Hb_000671_030 |
0.0790729784 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 6 |
Hb_007919_110 |
0.0798504056 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 7 |
Hb_004724_390 |
0.0832563221 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 8 |
Hb_000805_210 |
0.0838577101 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 9 |
Hb_002045_170 |
0.0887404802 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 10 |
Hb_006120_050 |
0.0911772797 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 11 |
Hb_000300_550 |
0.091474492 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_003777_130 |
0.0919945517 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 13 |
Hb_000054_040 |
0.0922182142 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 14 |
Hb_006022_010 |
0.094123895 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 15 |
Hb_003449_100 |
0.0956733341 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 16 |
Hb_000958_080 |
0.095709825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001250_050 |
0.0959840217 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000622_340 |
0.0964523926 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 19 |
Hb_000258_220 |
0.098315701 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000317_110 |
0.0983327183 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |