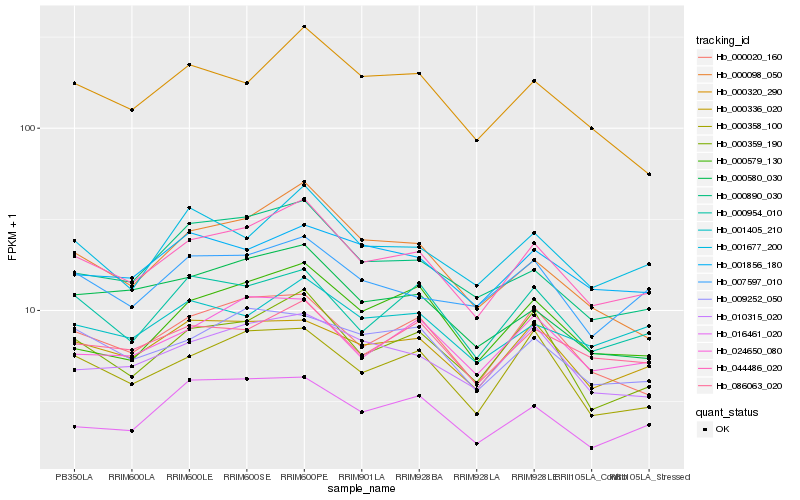
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044486_020 |
0.0 |
- |
- |
CASTOR protein [Glycine max] |
| 2 |
Hb_000359_190 |
0.0678216072 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 3 |
Hb_000020_160 |
0.0718126301 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 4 |
Hb_001677_200 |
0.0746937248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000890_030 |
0.0762711537 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 6 |
Hb_010315_020 |
0.0767831335 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 7 |
Hb_086063_020 |
0.0770487834 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000358_100 |
0.0782026154 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 9 |
Hb_000336_020 |
0.0787748283 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 10 |
Hb_000954_010 |
0.0790721152 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 11 |
Hb_024650_080 |
0.0794567387 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 12 |
Hb_007597_010 |
0.0817390395 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 13 |
Hb_000320_290 |
0.0819035085 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 14 |
Hb_009252_050 |
0.0832585525 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000580_030 |
0.083267812 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 16 |
Hb_000098_050 |
0.0833297231 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 17 |
Hb_000579_130 |
0.084170388 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_016461_020 |
0.0853606799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001405_210 |
0.08610035 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_001856_180 |
0.0864996477 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |