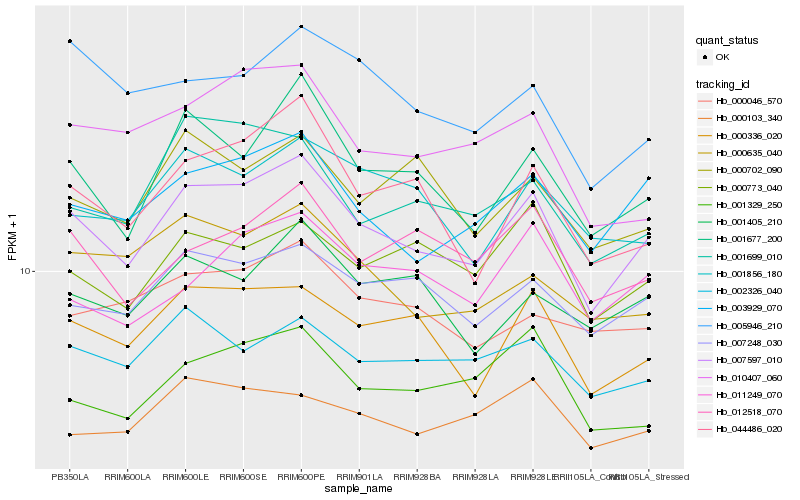
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007597_010 |
0.0 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 2 |
Hb_003929_070 |
0.0696250411 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 3 |
Hb_000336_020 |
0.0723033508 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 4 |
Hb_001677_200 |
0.0730764901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007248_030 |
0.0765646182 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 6 |
Hb_044486_020 |
0.0817390395 |
- |
- |
CASTOR protein [Glycine max] |
| 7 |
Hb_002326_040 |
0.0864780289 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 8 |
Hb_012518_070 |
0.088118202 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001405_210 |
0.0881372311 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 10 |
Hb_001856_180 |
0.089802391 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_000635_040 |
0.0899030498 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_001329_250 |
0.0908515866 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010407_060 |
0.0909908345 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 14 |
Hb_001699_010 |
0.0912172125 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 15 |
Hb_000773_040 |
0.0914375957 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000702_090 |
0.0929243232 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 17 |
Hb_011249_070 |
0.0930691676 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 18 |
Hb_005946_210 |
0.0930794795 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 19 |
Hb_000046_570 |
0.0933111007 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 20 |
Hb_000103_340 |
0.093694567 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |