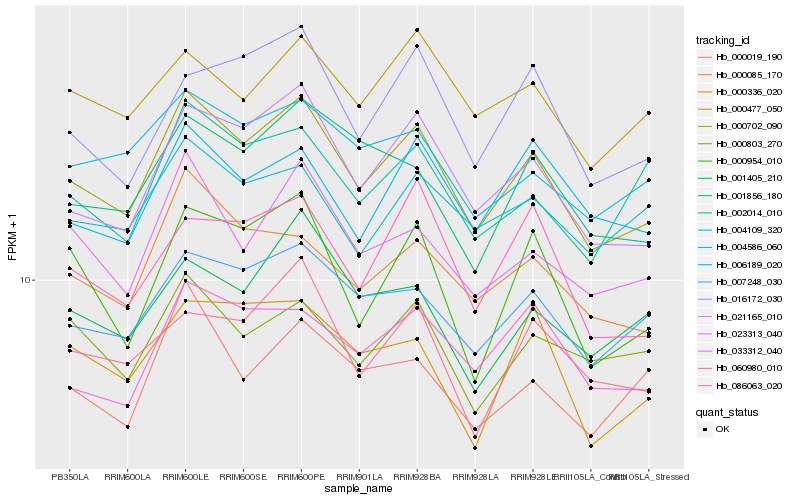
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000702_090 |
0.0 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 2 |
Hb_021165_010 |
0.0414976166 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_007248_030 |
0.0539331445 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 4 |
Hb_023313_040 |
0.0567689487 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000085_170 |
0.0573949391 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004586_060 |
0.0615995543 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 7 |
Hb_016172_030 |
0.0616635273 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 8 |
Hb_033312_040 |
0.0631006681 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_086063_020 |
0.0643974854 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 10 |
Hb_001856_180 |
0.0652768075 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_060980_010 |
0.065407716 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 12 |
Hb_000477_050 |
0.0659677489 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 13 |
Hb_002014_010 |
0.0661244097 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000954_010 |
0.0664740732 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 15 |
Hb_006189_020 |
0.0674738964 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 16 |
Hb_001405_210 |
0.0681157476 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 17 |
Hb_000803_270 |
0.0681936574 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 18 |
Hb_004109_320 |
0.0682070328 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 19 |
Hb_000336_020 |
0.0688338993 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 20 |
Hb_000019_190 |
0.0690755731 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |