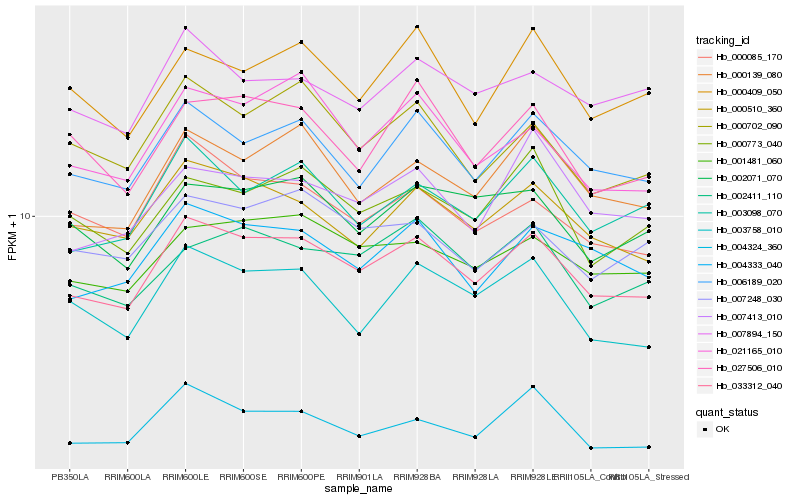
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_040 |
0.0 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_006189_020 |
0.0480705354 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 3 |
Hb_000510_360 |
0.0510091658 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 4 |
Hb_001481_060 |
0.0527673839 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000139_080 |
0.0540352996 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004333_040 |
0.0546736761 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 7 |
Hb_003098_070 |
0.0580042736 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 8 |
Hb_000085_170 |
0.0608142626 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003758_010 |
0.0610618853 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002411_110 |
0.062258088 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 11 |
Hb_021165_010 |
0.0626497963 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000702_090 |
0.0631006681 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 13 |
Hb_000773_040 |
0.0637261502 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002071_070 |
0.0639470359 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007894_150 |
0.0645534508 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 16 |
Hb_004324_360 |
0.0657170958 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007248_030 |
0.0657196303 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 18 |
Hb_007413_010 |
0.0666912678 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 19 |
Hb_000409_050 |
0.0667499711 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 20 |
Hb_027506_010 |
0.0668502234 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |