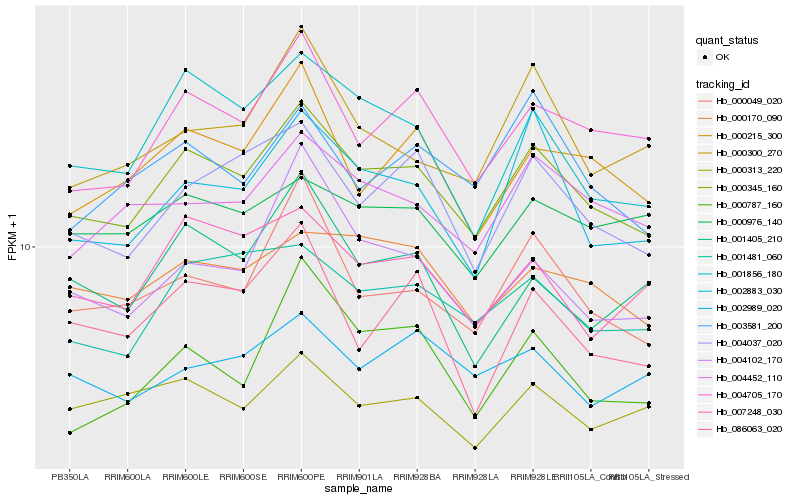
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_160 |
0.0 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 2 |
Hb_004037_020 |
0.0509394344 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004705_170 |
0.0516698887 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 4 |
Hb_001481_060 |
0.0518788599 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000049_020 |
0.053692412 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000170_090 |
0.054720482 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 7 |
Hb_004452_110 |
0.0552728269 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001856_180 |
0.0557662621 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 9 |
Hb_002883_030 |
0.0557854273 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 10 |
Hb_000215_300 |
0.057229945 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_007248_030 |
0.0588890308 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 12 |
Hb_000976_140 |
0.0625944686 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003581_200 |
0.063130839 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 14 |
Hb_000787_160 |
0.0634739038 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000313_220 |
0.0635109935 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004102_170 |
0.0639483386 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 17 |
Hb_000300_270 |
0.0642968555 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_002989_020 |
0.066143774 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001405_210 |
0.0680188906 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_086063_020 |
0.0683370933 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |