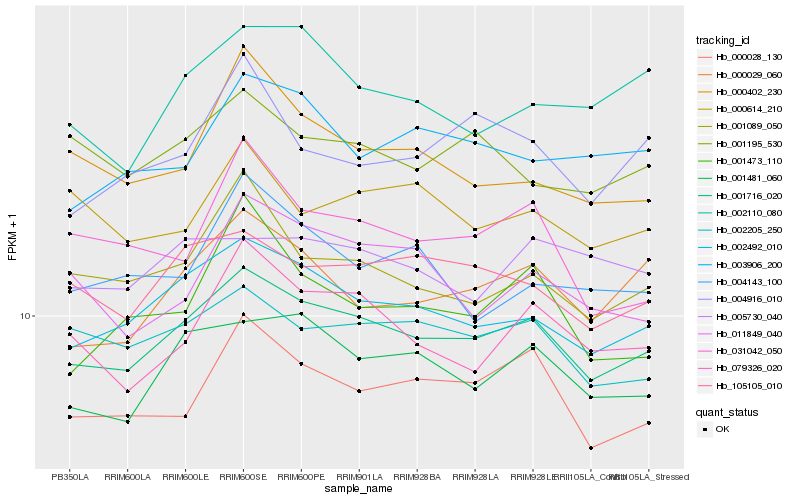
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001716_020 |
0.0 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 2 |
Hb_002492_010 |
0.0364425602 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 3 |
Hb_001481_060 |
0.0465009725 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_105105_010 |
0.0492518249 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 5 |
Hb_002205_250 |
0.0520174098 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000028_130 |
0.0520853981 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000029_060 |
0.055667779 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 8 |
Hb_001089_050 |
0.0557058086 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_079326_020 |
0.057876227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001195_530 |
0.0615161519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003906_200 |
0.0626603698 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 12 |
Hb_005730_040 |
0.064891932 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 13 |
Hb_002110_080 |
0.0651830352 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 14 |
Hb_000614_210 |
0.0653506934 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004916_010 |
0.0666626325 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 16 |
Hb_011849_040 |
0.0669521332 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_031042_050 |
0.0676057024 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 18 |
Hb_000402_230 |
0.0677864491 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001473_110 |
0.0683355172 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 20 |
Hb_004143_100 |
0.0683975788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |