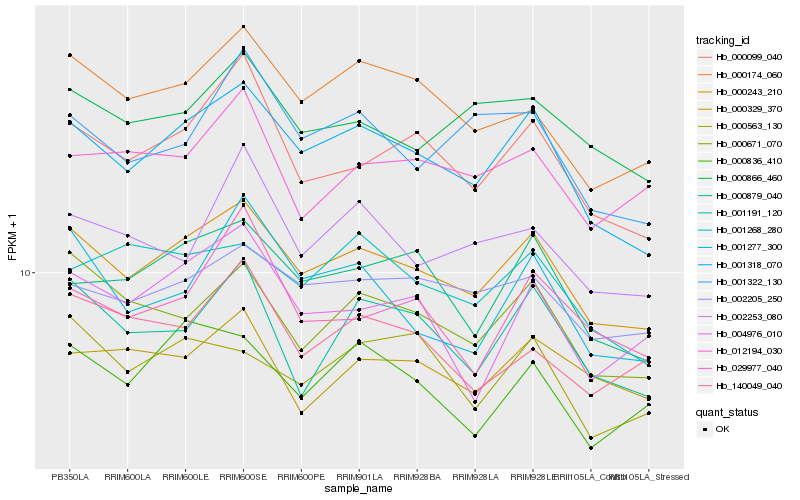
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_210 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_001318_070 |
0.0282058146 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000099_040 |
0.0511813778 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000174_060 |
0.0518726676 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_000671_070 |
0.052478951 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 6 |
Hb_002205_250 |
0.0525886065 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000879_040 |
0.0563180717 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 8 |
Hb_000866_460 |
0.058064968 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000836_410 |
0.0618067894 |
- |
- |
sec10, putative [Ricinus communis] |
| 10 |
Hb_001191_120 |
0.0640084662 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001268_280 |
0.0652776878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001322_130 |
0.065362512 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 13 |
Hb_000563_130 |
0.0659642899 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 14 |
Hb_001277_300 |
0.0663574411 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 15 |
Hb_012194_030 |
0.0667590001 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000329_370 |
0.0667626261 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 17 |
Hb_004976_010 |
0.0668003364 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 18 |
Hb_029977_040 |
0.0677656622 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 19 |
Hb_140049_040 |
0.0678005896 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 20 |
Hb_002253_080 |
0.0678762651 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |