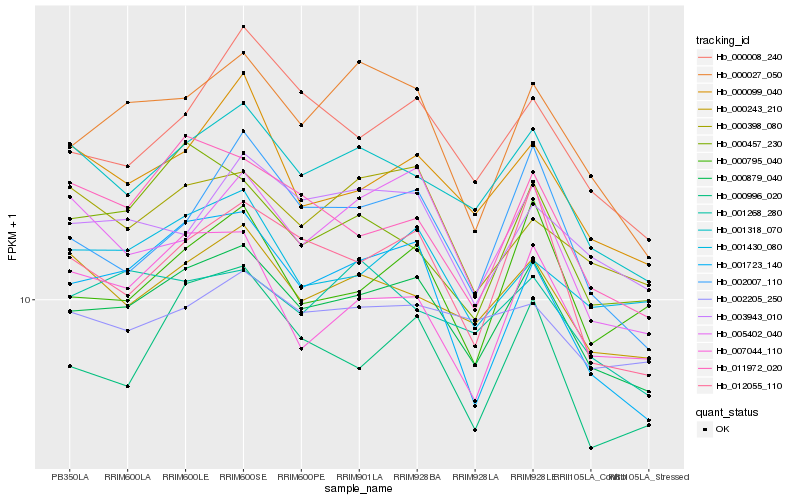
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_040 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 2 |
Hb_001318_070 |
0.0500191801 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000243_210 |
0.0563180717 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_012055_110 |
0.0598880869 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 5 |
Hb_000027_050 |
0.061374309 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000099_040 |
0.0617853738 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_001430_080 |
0.0623203773 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 8 |
Hb_003943_010 |
0.0636280871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000398_080 |
0.065412086 |
- |
- |
tip120, putative [Ricinus communis] |
| 10 |
Hb_000996_020 |
0.0658619517 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 11 |
Hb_000008_240 |
0.0660129307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000795_040 |
0.0660874549 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 13 |
Hb_001723_140 |
0.0661262428 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_001268_280 |
0.0667691876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005402_040 |
0.0678723929 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 16 |
Hb_011972_020 |
0.0682022563 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_007044_110 |
0.0682281233 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002205_250 |
0.068531332 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002007_110 |
0.0691763981 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000457_230 |
0.0702245039 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |