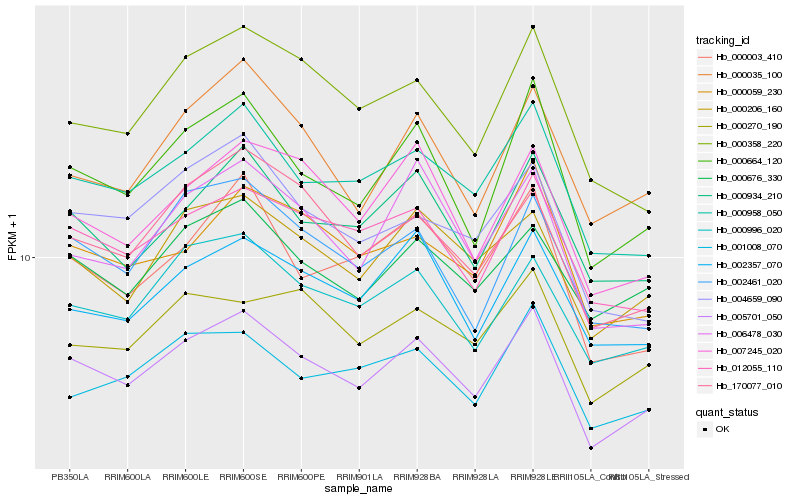
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005701_050 |
0.0 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 2 |
Hb_000664_120 |
0.0531494693 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000958_050 |
0.0545307179 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 4 |
Hb_012055_110 |
0.062049878 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 5 |
Hb_000996_020 |
0.0640559171 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 6 |
Hb_000934_210 |
0.064072391 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 7 |
Hb_000206_160 |
0.0642423824 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000676_330 |
0.0697251106 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_002461_020 |
0.0706856446 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007245_020 |
0.0712181212 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 11 |
Hb_002357_070 |
0.0723614063 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 12 |
Hb_000059_230 |
0.072588459 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 13 |
Hb_170077_010 |
0.0736538025 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000003_410 |
0.0750109707 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 15 |
Hb_004659_090 |
0.0752391296 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 16 |
Hb_000358_220 |
0.0753505177 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 17 |
Hb_000035_100 |
0.075765616 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_006478_030 |
0.0761891092 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 19 |
Hb_001008_070 |
0.0774253679 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 20 |
Hb_000270_190 |
0.0774464816 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |