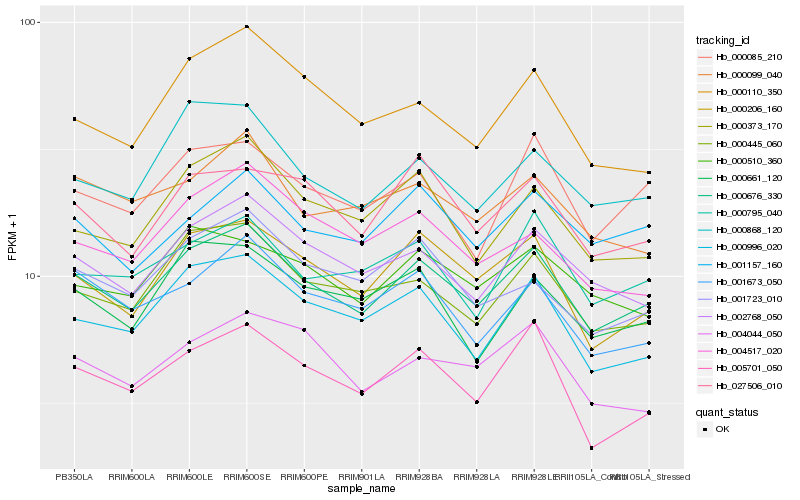
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_330 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000206_160 |
0.0545265816 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_002768_050 |
0.0579003328 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_000445_060 |
0.0588569005 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000996_020 |
0.0598358284 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 6 |
Hb_000868_120 |
0.0652998917 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 7 |
Hb_027506_010 |
0.0669048854 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 8 |
Hb_001157_160 |
0.0669290834 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000795_040 |
0.0676559883 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 10 |
Hb_000510_360 |
0.0678210538 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 11 |
Hb_001673_050 |
0.0678214713 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000085_210 |
0.0682304459 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 13 |
Hb_000373_170 |
0.0690407764 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 14 |
Hb_005701_050 |
0.0697251106 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 15 |
Hb_000099_040 |
0.0703205321 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_001723_010 |
0.0724138319 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 17 |
Hb_000661_120 |
0.072467186 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 18 |
Hb_004044_050 |
0.0737420641 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 19 |
Hb_000110_350 |
0.0751463859 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_004517_020 |
0.0754041978 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |