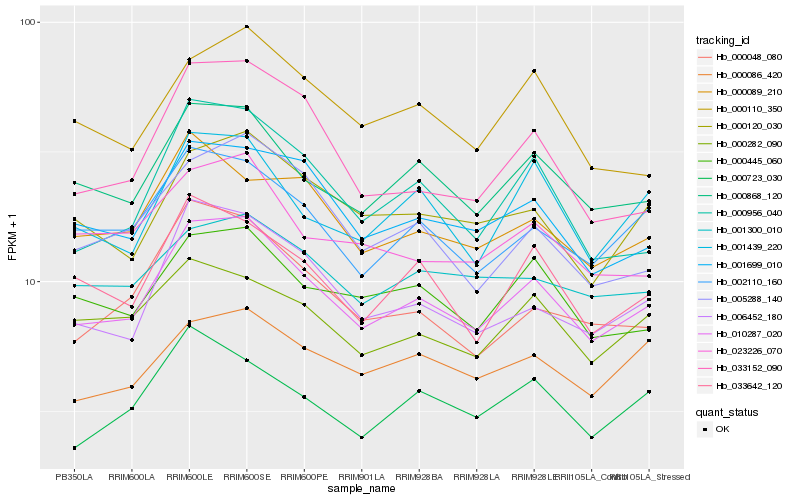
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010287_020 |
0.0 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 2 |
Hb_000868_120 |
0.0559066798 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 3 |
Hb_006452_180 |
0.0643582994 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 4 |
Hb_002110_160 |
0.0652851239 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001699_010 |
0.0655867223 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 6 |
Hb_033642_120 |
0.0705621267 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 7 |
Hb_000445_060 |
0.0709274101 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000956_040 |
0.0714825441 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 9 |
Hb_000120_030 |
0.0723076639 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001439_220 |
0.0749831019 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 11 |
Hb_005288_140 |
0.0755600286 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 12 |
Hb_000048_080 |
0.0766058357 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_023226_070 |
0.077378197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000282_090 |
0.0792922215 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.0796354343 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_000723_030 |
0.0797436787 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 17 |
Hb_033152_090 |
0.0804277347 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 18 |
Hb_000110_350 |
0.0804472246 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_001300_010 |
0.0808807814 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000086_420 |
0.0809962324 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |