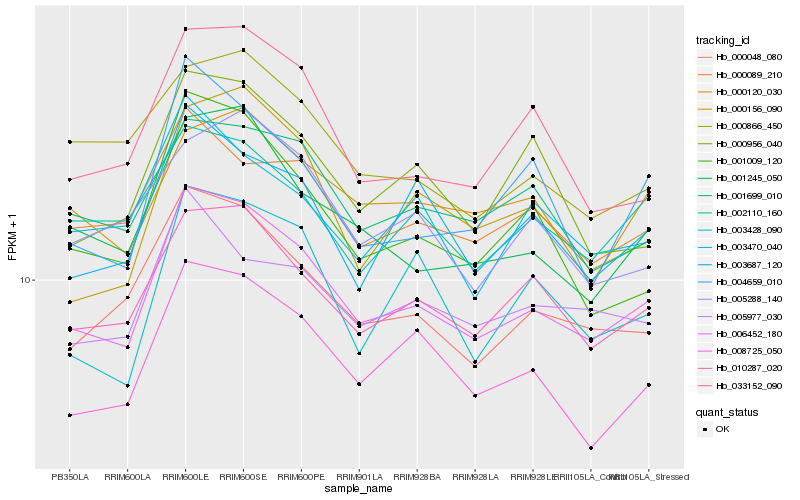
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006452_180 |
0.0 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 2 |
Hb_010287_020 |
0.0643582994 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 3 |
Hb_000048_080 |
0.0686896657 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008725_050 |
0.0731324978 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_001245_050 |
0.0733519466 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 6 |
Hb_001009_120 |
0.0812836926 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000089_210 |
0.0813740861 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_001699_010 |
0.0816971049 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 9 |
Hb_003687_120 |
0.0827585785 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 10 |
Hb_033152_090 |
0.0839669233 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 11 |
Hb_005977_030 |
0.0846616458 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 12 |
Hb_000120_030 |
0.0863551524 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002110_160 |
0.086668943 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000156_090 |
0.086791275 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004659_010 |
0.0886991988 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |
| 16 |
Hb_003428_090 |
0.0902334841 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 17 |
Hb_000956_040 |
0.0917670471 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 18 |
Hb_005288_140 |
0.0921043539 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 19 |
Hb_003470_040 |
0.0927721156 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000866_450 |
0.0934241354 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |