| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
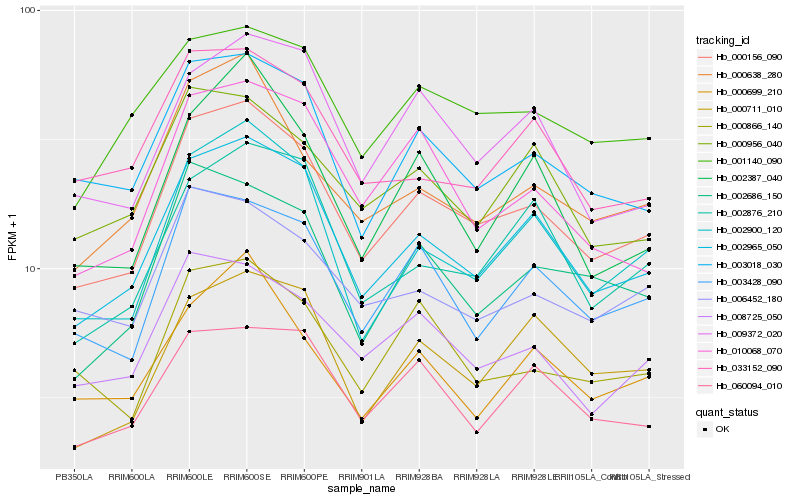
Hb_000156_090 |
0.0 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002965_050 |
0.0499939983 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 3 |
Hb_003018_030 |
0.0741901604 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 4 |
Hb_002900_120 |
0.0742782066 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 5 |
Hb_003428_090 |
0.075608902 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 6 |
Hb_008725_050 |
0.0756936534 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_000638_280 |
0.0803530211 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 8 |
Hb_002686_150 |
0.0836682007 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_001140_090 |
0.0848857276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000866_140 |
0.085641748 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 11 |
Hb_006452_180 |
0.086791275 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 12 |
Hb_000956_040 |
0.0882293514 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 13 |
Hb_009372_020 |
0.0904516222 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 14 |
Hb_060094_010 |
0.0911845535 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010068_070 |
0.0914746263 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002876_210 |
0.0920701637 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 17 |
Hb_033152_090 |
0.093517906 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 18 |
Hb_000699_210 |
0.0945497859 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_000711_010 |
0.0951343879 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 20 |
Hb_002387_040 |
0.0971591693 |
- |
- |
protein transporter, putative [Ricinus communis] |