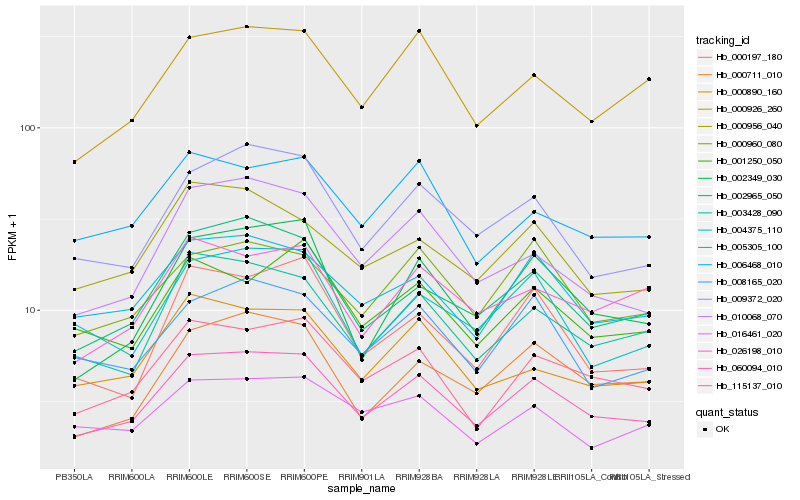
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060094_010 |
0.0 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 2 |
Hb_115137_010 |
0.0650666527 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002965_050 |
0.0700898743 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 4 |
Hb_005305_100 |
0.0713765784 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003428_090 |
0.0719709381 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 6 |
Hb_009372_020 |
0.0730395554 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 7 |
Hb_010068_070 |
0.0811121581 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_002349_030 |
0.0822363041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000926_260 |
0.0826611097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000956_040 |
0.083089715 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 11 |
Hb_008165_020 |
0.0831828341 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006468_010 |
0.0851039485 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000890_160 |
0.0853190902 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 14 |
Hb_000711_010 |
0.0855656513 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000960_080 |
0.0874570798 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_026198_010 |
0.0901042212 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001250_050 |
0.0903897194 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004375_110 |
0.0905632473 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 19 |
Hb_016461_020 |
0.0905812198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000197_180 |
0.0910835814 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |