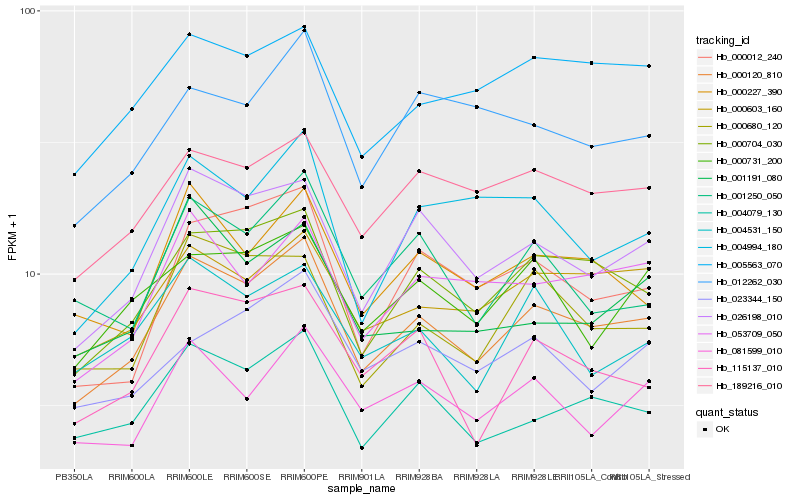
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_810 |
0.0 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 2 |
Hb_000704_030 |
0.0626298055 |
- |
- |
PREDICTED: probable aquaporin SIP2-1 [Cicer arietinum] |
| 3 |
Hb_026198_010 |
0.0670440087 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004079_130 |
0.0735842593 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 5 |
Hb_000603_160 |
0.0852251842 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_189216_010 |
0.0857003498 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000680_120 |
0.0868661083 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 8 |
Hb_000227_390 |
0.0872560606 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_053709_050 |
0.0893361257 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_005563_070 |
0.0920463395 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 11 |
Hb_115137_010 |
0.0931617693 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000731_200 |
0.0940962807 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 13 |
Hb_081599_010 |
0.0941261638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000012_240 |
0.0963987676 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004531_150 |
0.0965074922 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001250_050 |
0.0975430109 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012262_030 |
0.0987095368 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004994_180 |
0.0987234855 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 19 |
Hb_001191_080 |
0.1017620494 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_023344_150 |
0.1033223715 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |