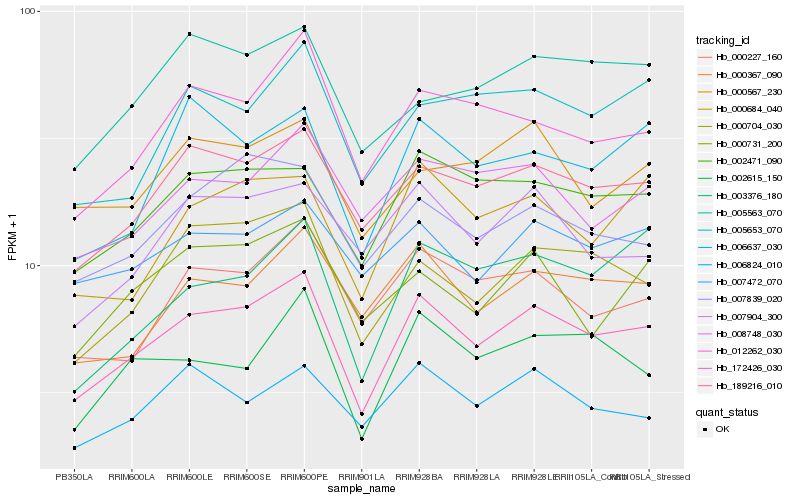
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_030 |
0.0 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 2 |
Hb_189216_010 |
0.0731023008 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002471_090 |
0.0787200781 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 4 |
Hb_000704_030 |
0.0819442794 |
- |
- |
PREDICTED: probable aquaporin SIP2-1 [Cicer arietinum] |
| 5 |
Hb_012262_030 |
0.0880534906 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000367_090 |
0.0889997513 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 7 |
Hb_000227_160 |
0.091750478 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 8 |
Hb_000684_040 |
0.0919829472 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 9 |
Hb_002615_150 |
0.0920113621 |
- |
- |
PREDICTED: crt homolog 1-like [Jatropha curcas] |
| 10 |
Hb_007472_070 |
0.0937397606 |
- |
- |
cir, putative [Ricinus communis] |
| 11 |
Hb_007839_020 |
0.0938908175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006637_030 |
0.0940523757 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 13 |
Hb_008748_030 |
0.095249437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006824_010 |
0.0960341395 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 15 |
Hb_005653_070 |
0.0978148248 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 16 |
Hb_007904_300 |
0.0980596114 |
- |
- |
copine, putative [Ricinus communis] |
| 17 |
Hb_003376_180 |
0.0989476591 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 18 |
Hb_000567_230 |
0.0989698483 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 19 |
Hb_000731_200 |
0.0992111253 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005563_070 |
0.0992277081 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |