| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
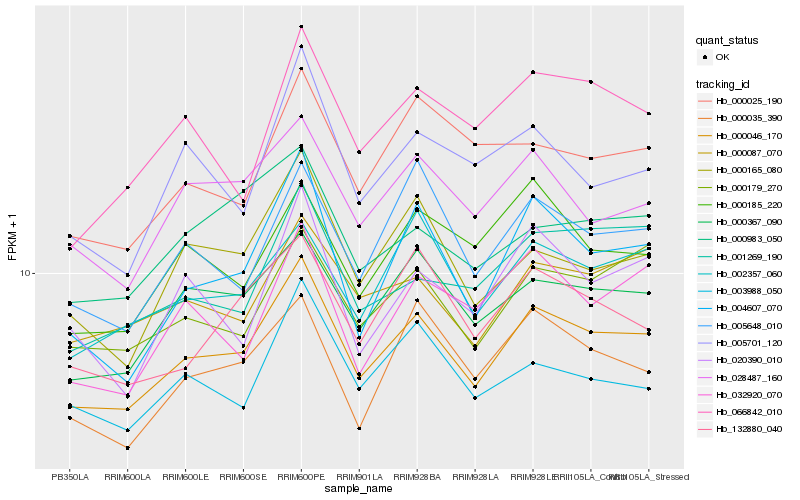
Hb_000046_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000367_090 |
0.068259911 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 3 |
Hb_004607_070 |
0.0764377629 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 4 |
Hb_000179_270 |
0.0798219498 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 5 |
Hb_028487_160 |
0.080381842 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 6 |
Hb_000087_070 |
0.0809057991 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 7 |
Hb_000165_080 |
0.0825636966 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002357_060 |
0.0837877964 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 9 |
Hb_005648_010 |
0.0856339321 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 10 |
Hb_132880_040 |
0.0867878109 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_003988_050 |
0.0873449233 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 12 |
Hb_000983_050 |
0.0880696867 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000035_390 |
0.088336709 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 14 |
Hb_001269_190 |
0.0913106914 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 15 |
Hb_066842_010 |
0.0924796567 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000025_190 |
0.0931809582 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 17 |
Hb_020390_010 |
0.0938412521 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 18 |
Hb_000185_220 |
0.0939036574 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 19 |
Hb_032920_070 |
0.0942520204 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 20 |
Hb_005701_120 |
0.095373639 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |