| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
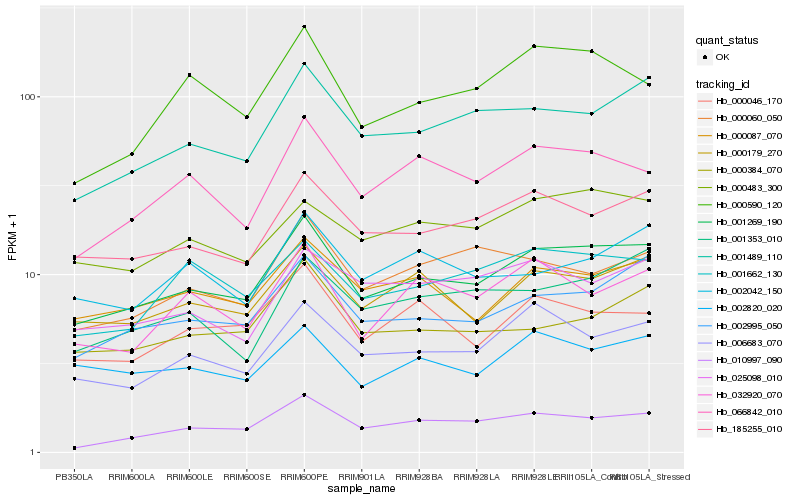
Hb_001269_190 |
0.0 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 2 |
Hb_185255_010 |
0.0699106309 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 3 |
Hb_002995_050 |
0.0737528376 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001489_110 |
0.075591786 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 5 |
Hb_000179_270 |
0.078356621 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 6 |
Hb_006683_070 |
0.0791712676 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 7 |
Hb_000087_070 |
0.079798755 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 8 |
Hb_066842_010 |
0.0859904969 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000483_300 |
0.0889660446 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 10 |
Hb_000046_170 |
0.0913106914 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000384_070 |
0.0927491733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001662_130 |
0.0931483053 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002820_020 |
0.0935797104 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 14 |
Hb_000590_120 |
0.096223647 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 15 |
Hb_010997_090 |
0.0977406314 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 16 |
Hb_032920_070 |
0.0977733209 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 17 |
Hb_000060_050 |
0.099265734 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 18 |
Hb_001353_010 |
0.0995585183 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002042_150 |
0.1008878642 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 20 |
Hb_025098_010 |
0.1013777435 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |