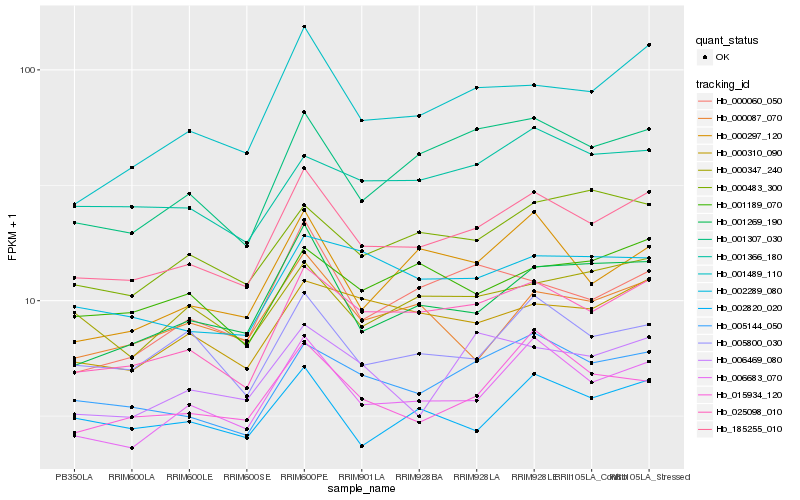
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185255_010 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 2 |
Hb_025098_010 |
0.0526911885 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_006683_070 |
0.060372966 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 4 |
Hb_005144_050 |
0.0689314146 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 5 |
Hb_001269_190 |
0.0699106309 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 6 |
Hb_001489_110 |
0.0711990781 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 7 |
Hb_001307_030 |
0.0742149128 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000310_090 |
0.0815697775 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005800_030 |
0.0834787917 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 10 |
Hb_015934_120 |
0.0846936291 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 11 |
Hb_000297_120 |
0.0849567997 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 12 |
Hb_002820_020 |
0.085604114 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 13 |
Hb_000347_240 |
0.0860922713 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 14 |
Hb_001366_180 |
0.0861892715 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000060_050 |
0.0874354063 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 16 |
Hb_000483_300 |
0.0885596179 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 17 |
Hb_002289_080 |
0.0888461953 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 18 |
Hb_001189_070 |
0.0894341305 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 19 |
Hb_006469_080 |
0.0898153739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000087_070 |
0.0899640156 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |