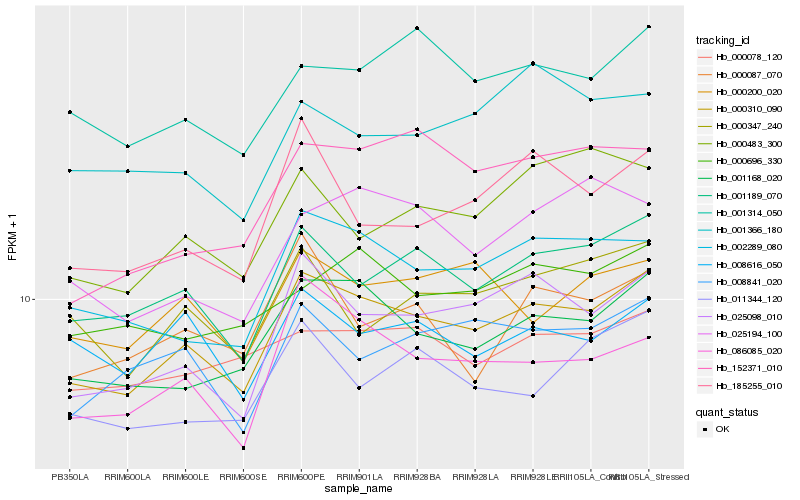
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000310_090 |
0.0 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001168_020 |
0.0581008785 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 3 |
Hb_001189_070 |
0.0604379424 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 4 |
Hb_025098_010 |
0.0640586181 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_002289_080 |
0.0673199973 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 6 |
Hb_086085_020 |
0.0724986901 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 7 |
Hb_001314_050 |
0.077607666 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit alpha-like [Jatropha curcas] |
| 8 |
Hb_008841_020 |
0.0777097674 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000200_020 |
0.07966713 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 10 |
Hb_185255_010 |
0.0815697775 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 11 |
Hb_000078_120 |
0.0820400814 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 12 |
Hb_000347_240 |
0.0831809239 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 13 |
Hb_000696_330 |
0.0837755785 |
- |
- |
PREDICTED: uncharacterized protein LOC105647254 [Jatropha curcas] |
| 14 |
Hb_011344_120 |
0.0852505024 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 15 |
Hb_000483_300 |
0.0860540112 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 16 |
Hb_025194_100 |
0.0863664944 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 17 |
Hb_152371_010 |
0.0880141442 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |
| 18 |
Hb_008616_050 |
0.0881961597 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 19 |
Hb_000087_070 |
0.0899154008 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 20 |
Hb_001366_180 |
0.0902117661 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |