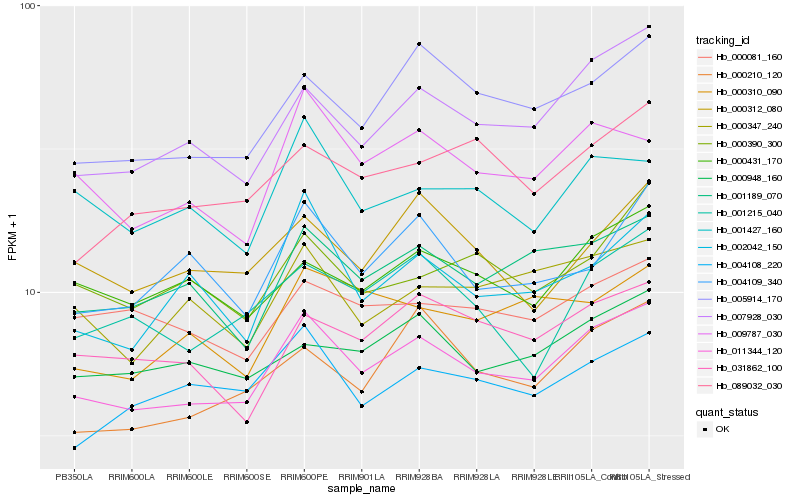
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 2 |
Hb_007928_030 |
0.0632534732 |
- |
- |
PREDICTED: dual specificity phosphatase Cdc25 [Jatropha curcas] |
| 3 |
Hb_005914_170 |
0.0665605688 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 4 |
Hb_001189_070 |
0.0696356857 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 5 |
Hb_009787_030 |
0.0757014788 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 6 |
Hb_004108_220 |
0.0801733333 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000347_240 |
0.0805460484 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 8 |
Hb_000431_170 |
0.080617795 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 9 |
Hb_000948_160 |
0.081065509 |
- |
- |
PREDICTED: 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000210_120 |
0.0821973542 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 11 |
Hb_031862_100 |
0.0822595334 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002042_150 |
0.0824057811 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 13 |
Hb_001215_040 |
0.0835236174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_089032_030 |
0.0846697979 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000310_090 |
0.0852505024 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000312_080 |
0.0853087033 |
- |
- |
PREDICTED: transmembrane protein 97-like [Jatropha curcas] |
| 17 |
Hb_000081_160 |
0.0855588946 |
- |
- |
PREDICTED: uncharacterized protein At4g17910 [Jatropha curcas] |
| 18 |
Hb_001427_160 |
0.0857413265 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_004109_340 |
0.0862705896 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |
| 20 |
Hb_000390_300 |
0.0877549637 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |