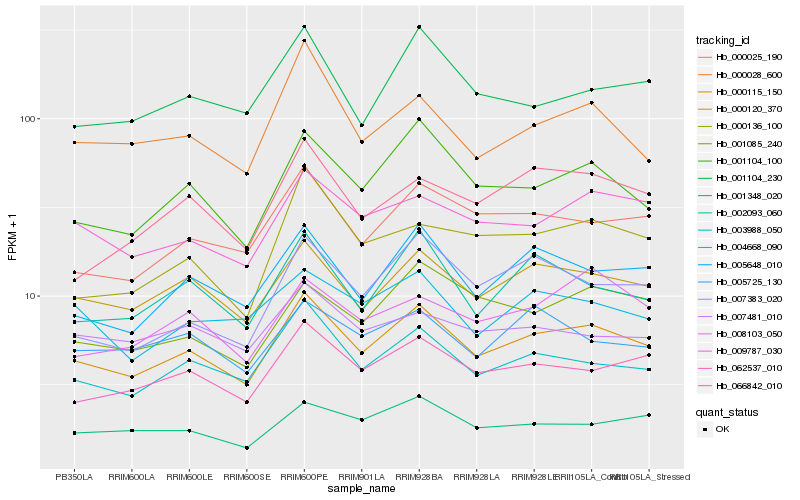
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_150 |
0.0 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 2 |
Hb_000120_370 |
0.0610137267 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 3 |
Hb_003988_050 |
0.0686177038 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 4 |
Hb_062537_010 |
0.0737500874 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 5 |
Hb_001104_100 |
0.0748708026 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 6 |
Hb_005648_010 |
0.0770398641 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_190 |
0.0801991992 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_009787_030 |
0.081402197 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 9 |
Hb_002093_060 |
0.0845515787 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_001348_020 |
0.0886635934 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004668_090 |
0.0942292867 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 12 |
Hb_001104_230 |
0.0951492762 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 13 |
Hb_005725_130 |
0.0966613648 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_001085_240 |
0.0973137091 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007383_020 |
0.0976799193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000028_600 |
0.0988032043 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 17 |
Hb_007481_010 |
0.0999526836 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 18 |
Hb_066842_010 |
0.099976225 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_008103_050 |
0.1010132786 |
- |
- |
hypothetical protein JCGZ_07422 [Jatropha curcas] |
| 20 |
Hb_000136_100 |
0.1016173571 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |