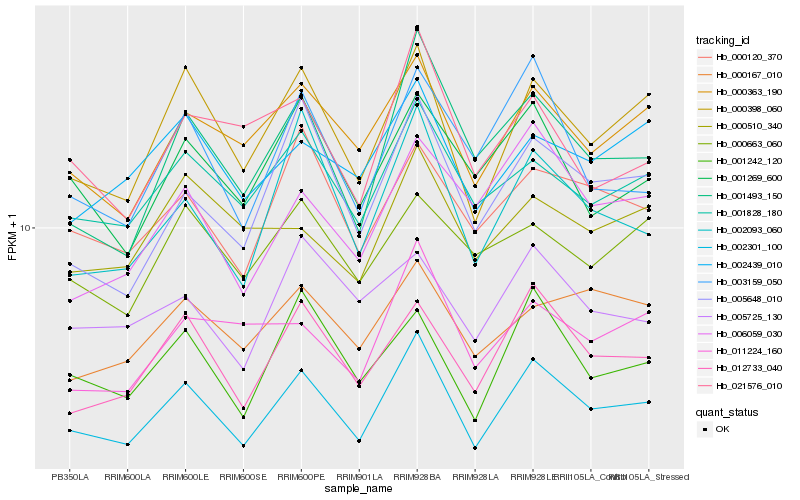
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 2 |
Hb_000398_060 |
0.0580172826 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 3 |
Hb_012733_040 |
0.0666875472 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 4 |
Hb_001828_180 |
0.0694849752 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 5 |
Hb_000363_190 |
0.0699281981 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001242_120 |
0.0756587048 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 7 |
Hb_001493_150 |
0.0766277477 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 8 |
Hb_000120_370 |
0.0781365476 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 9 |
Hb_002093_060 |
0.0797086409 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_005648_010 |
0.0822649136 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 11 |
Hb_001269_600 |
0.0823998902 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_003159_050 |
0.0847374114 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_002439_010 |
0.0859547634 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 14 |
Hb_000167_010 |
0.0878086603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005725_130 |
0.0901555918 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_011224_160 |
0.0901629879 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 17 |
Hb_021576_010 |
0.0915307148 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 18 |
Hb_000663_060 |
0.0920075729 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 19 |
Hb_000510_340 |
0.0920228837 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 20 |
Hb_006059_030 |
0.0926093881 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |