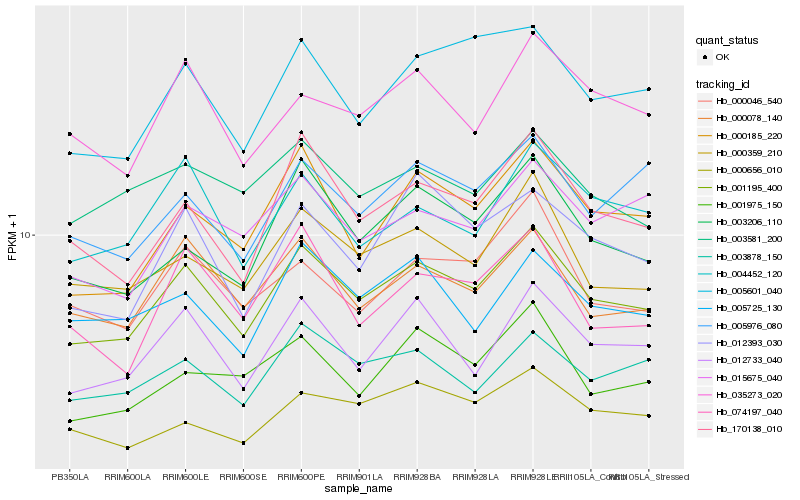
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_400 |
0.0 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 2 |
Hb_000078_140 |
0.0619800791 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 3 |
Hb_170138_010 |
0.0619922115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012733_040 |
0.0651724123 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 5 |
Hb_000359_210 |
0.0656201612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000185_220 |
0.0673160486 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 7 |
Hb_003206_110 |
0.0686097224 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 8 |
Hb_005601_040 |
0.0702886402 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_003878_150 |
0.0726324169 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_004452_120 |
0.0747900202 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 11 |
Hb_012393_030 |
0.075564033 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 12 |
Hb_035273_020 |
0.0759741976 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000046_540 |
0.0768848503 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 14 |
Hb_005725_130 |
0.0771010596 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_015675_040 |
0.0771019658 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 16 |
Hb_005976_080 |
0.0773575362 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 17 |
Hb_000656_010 |
0.0778498689 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001975_150 |
0.0783382015 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 19 |
Hb_003581_200 |
0.0784999117 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 20 |
Hb_074197_040 |
0.0785635289 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |