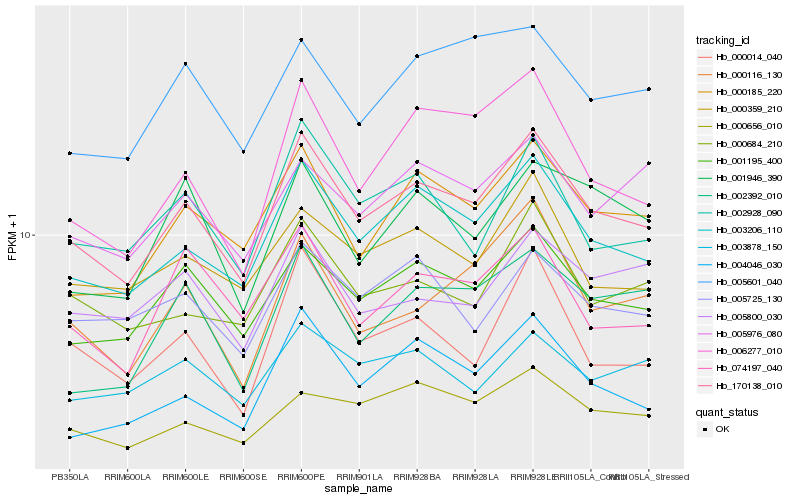
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170138_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003206_110 |
0.045883645 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 3 |
Hb_001195_400 |
0.0619922115 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 4 |
Hb_000185_220 |
0.0727418288 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 5 |
Hb_004046_030 |
0.0778877091 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 6 |
Hb_000359_210 |
0.0782359998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000684_210 |
0.0803626968 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 8 |
Hb_002928_090 |
0.081696147 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 9 |
Hb_006277_010 |
0.0817118931 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 10 |
Hb_005725_130 |
0.0850753235 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_000656_010 |
0.0851193227 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000014_040 |
0.0852666223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_074197_040 |
0.0857918821 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 14 |
Hb_005976_080 |
0.0887718182 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 15 |
Hb_005601_040 |
0.0889046217 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000116_130 |
0.0889389299 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 17 |
Hb_005800_030 |
0.0903945334 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 18 |
Hb_003878_150 |
0.0913922511 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 19 |
Hb_001946_390 |
0.0933869796 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 20 |
Hb_002392_010 |
0.0935513329 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |