| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
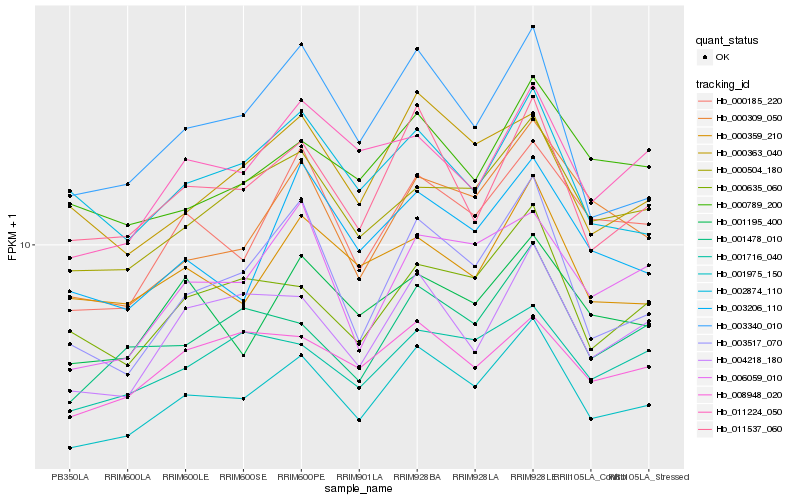
Hb_001975_150 |
0.0 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 2 |
Hb_011537_060 |
0.0619869139 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 3 |
Hb_000504_180 |
0.0670548168 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_000185_220 |
0.0697724867 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 5 |
Hb_004218_180 |
0.0733291719 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 6 |
Hb_000635_060 |
0.0758880816 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 7 |
Hb_003517_070 |
0.0759258635 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 8 |
Hb_001478_010 |
0.0775861691 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001195_400 |
0.0783382015 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 10 |
Hb_000359_210 |
0.078708096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003340_010 |
0.0810864426 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 12 |
Hb_000309_050 |
0.0811785174 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003206_110 |
0.0814852307 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 14 |
Hb_000789_200 |
0.0815156035 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 15 |
Hb_006059_010 |
0.0821932485 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 16 |
Hb_002874_110 |
0.0827675136 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 17 |
Hb_011224_050 |
0.0833796997 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 18 |
Hb_001716_040 |
0.0841448391 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 19 |
Hb_000363_040 |
0.0844799986 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 20 |
Hb_008948_020 |
0.0849906575 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |