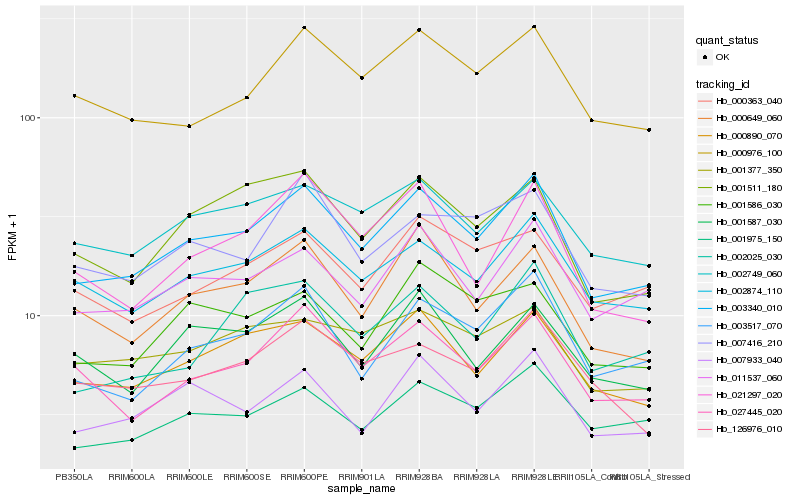
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003340_010 |
0.0 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 2 |
Hb_000890_070 |
0.0665644263 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002874_110 |
0.0721462285 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 4 |
Hb_003517_070 |
0.0730150015 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 5 |
Hb_011537_060 |
0.0773424091 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 6 |
Hb_007933_040 |
0.0792728198 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000649_060 |
0.0796920771 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 8 |
Hb_002749_060 |
0.0806669424 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_001975_150 |
0.0810864426 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 10 |
Hb_027445_020 |
0.0811519593 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 11 |
Hb_001511_180 |
0.0814653596 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 12 |
Hb_000976_100 |
0.0859324869 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 13 |
Hb_007416_210 |
0.0888326739 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 14 |
Hb_001586_030 |
0.0898098564 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 15 |
Hb_001587_030 |
0.0898819795 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 16 |
Hb_002025_030 |
0.091629971 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000363_040 |
0.0919103171 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 18 |
Hb_001377_350 |
0.0920011865 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 19 |
Hb_126976_010 |
0.0920430633 |
- |
- |
- |
| 20 |
Hb_021297_020 |
0.092050725 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |