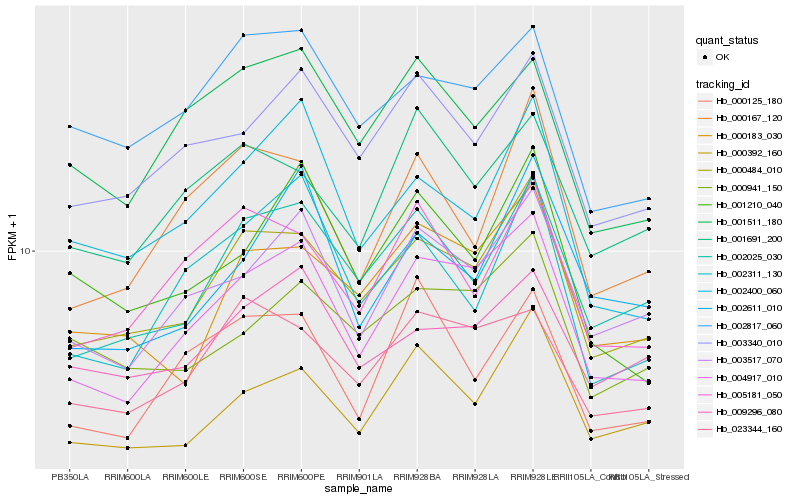
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004917_010 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 2 |
Hb_003517_070 |
0.0750898672 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 3 |
Hb_000484_010 |
0.0825226509 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 4 |
Hb_003340_010 |
0.0999459499 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 5 |
Hb_002025_030 |
0.104051578 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_023344_160 |
0.106950682 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 7 |
Hb_000392_160 |
0.1107037889 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000183_030 |
0.1113790846 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0291141 [Populus euphratica] |
| 9 |
Hb_001511_180 |
0.1138500567 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 10 |
Hb_002400_060 |
0.1147649638 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 11 |
Hb_000941_150 |
0.1175907354 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001210_040 |
0.1178434272 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 13 |
Hb_002311_130 |
0.1182225223 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000125_180 |
0.1183941721 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 15 |
Hb_002817_060 |
0.1184522676 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 16 |
Hb_002611_010 |
0.1186365817 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_005181_050 |
0.1197916904 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 18 |
Hb_009296_080 |
0.1211024063 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000167_120 |
0.1225713224 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_001691_200 |
0.1225930128 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |