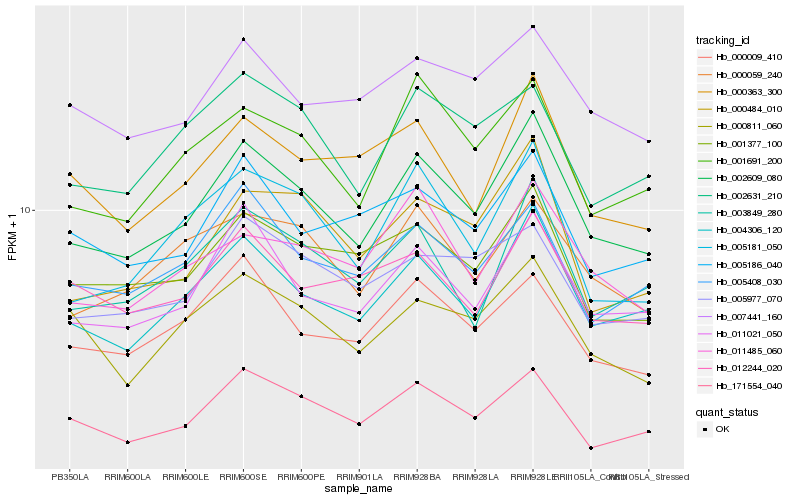
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_080 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_004306_120 |
0.0530583403 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_011021_050 |
0.0645964104 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001691_200 |
0.0758622205 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 5 |
Hb_001377_100 |
0.0793750086 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003849_280 |
0.0811894156 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 7 |
Hb_005181_050 |
0.081251228 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000009_410 |
0.0822263518 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 9 |
Hb_000059_240 |
0.08318385 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 10 |
Hb_011485_060 |
0.0841599574 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 11 |
Hb_005408_030 |
0.0842262408 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 12 |
Hb_000363_300 |
0.0846306167 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000484_010 |
0.0853024531 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 14 |
Hb_171554_040 |
0.0862525538 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 15 |
Hb_005186_040 |
0.0872559405 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005977_070 |
0.0876416124 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007441_160 |
0.0878675758 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002631_210 |
0.0887808594 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 19 |
Hb_012244_020 |
0.0899918444 |
- |
- |
calpain, putative [Ricinus communis] |
| 20 |
Hb_000811_060 |
0.0907807362 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |